
Avian metaavulavirus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
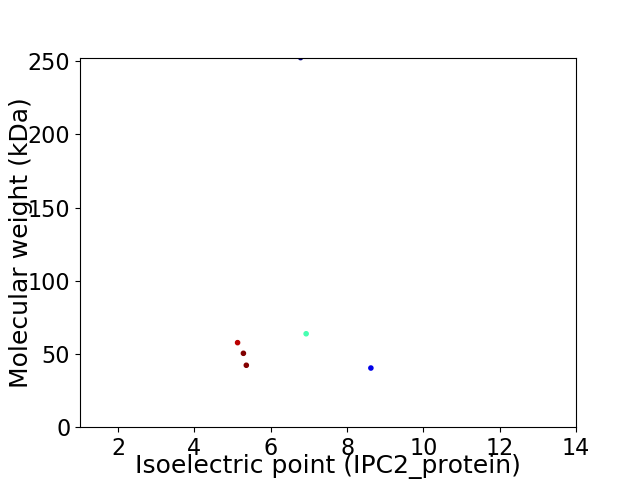
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q916F8|Q916F8_9MONO Hemagglutinin-neuraminidase OS=Avian metaavulavirus 2 OX=2560313 GN=HN PE=3 SV=1
MM1 pKa = 6.87NQALVILLVSFQLGVALDD19 pKa = 3.93NSVLAPIGVASAQEE33 pKa = 4.0WQLAAYY39 pKa = 6.49TTTLTGTIAVRR50 pKa = 11.84FIPVLPGNLSTCAQEE65 pKa = 4.48TLQEE69 pKa = 4.11YY70 pKa = 10.47NRR72 pKa = 11.84TVTNILGPLRR82 pKa = 11.84EE83 pKa = 4.36NLDD86 pKa = 3.68ALLSDD91 pKa = 4.23FDD93 pKa = 4.25KK94 pKa = 10.78PASRR98 pKa = 11.84FVGAIIGSVALGVATAAQITAAVALNQAQEE128 pKa = 3.82NARR131 pKa = 11.84NIWRR135 pKa = 11.84LKK137 pKa = 10.27EE138 pKa = 4.31SIKK141 pKa = 9.74KK142 pKa = 8.26TNEE145 pKa = 3.36AVLEE149 pKa = 4.42LKK151 pKa = 10.82DD152 pKa = 4.01GLATTAIALDD162 pKa = 3.89KK163 pKa = 10.27VQKK166 pKa = 10.5FINDD170 pKa = 4.08DD171 pKa = 4.22IIPQIKK177 pKa = 10.48DD178 pKa = 2.7IDD180 pKa = 4.17CQVVANKK187 pKa = 10.52LGVYY191 pKa = 10.44LSLYY195 pKa = 8.74LTEE198 pKa = 4.3LTTVFGSQITNPALSTLSYY217 pKa = 9.72QALYY221 pKa = 9.92SLCGGDD227 pKa = 3.27MGKK230 pKa = 8.33LTEE233 pKa = 5.08LIGVNAKK240 pKa = 10.31DD241 pKa = 3.4VGSLYY246 pKa = 10.33EE247 pKa = 4.26ANLITGQIVGYY258 pKa = 10.32DD259 pKa = 3.66PEE261 pKa = 4.29LQIILIQVSYY271 pKa = 10.28PSVSEE276 pKa = 4.1VTGVRR281 pKa = 11.84ATEE284 pKa = 4.01LVTVSVTTPKK294 pKa = 11.1GEE296 pKa = 4.01GQAIVPRR303 pKa = 11.84YY304 pKa = 7.88VAQSRR309 pKa = 11.84VLTEE313 pKa = 3.93EE314 pKa = 4.94LDD316 pKa = 3.61VSTCRR321 pKa = 11.84FSKK324 pKa = 7.66TTLYY328 pKa = 10.42CRR330 pKa = 11.84SILTRR335 pKa = 11.84PLPTLIASCLSGKK348 pKa = 10.3YY349 pKa = 9.82DD350 pKa = 3.52DD351 pKa = 4.82CQYY354 pKa = 8.24TTEE357 pKa = 4.23IGALSSRR364 pKa = 11.84FITVNGGVLANCRR377 pKa = 11.84AIVCKK382 pKa = 10.23CVSPPHH388 pKa = 7.16IIPQNDD394 pKa = 3.17IGSVTVIDD402 pKa = 3.88SSICKK407 pKa = 9.49EE408 pKa = 4.06VVLEE412 pKa = 4.16SVQLRR417 pKa = 11.84LEE419 pKa = 4.66GKK421 pKa = 10.18LSSQYY426 pKa = 10.48FSNVTIDD433 pKa = 3.86LSQITTSGSLDD444 pKa = 3.1ISSEE448 pKa = 3.63IGSINNTVNRR458 pKa = 11.84VDD460 pKa = 3.94EE461 pKa = 4.94LIKK464 pKa = 10.72EE465 pKa = 4.33SNEE468 pKa = 3.23WLNAVNPRR476 pKa = 11.84LVNNTSIIVLCVLAALIIVWLIALTVCFCYY506 pKa = 10.1SARR509 pKa = 11.84YY510 pKa = 8.12SAKK513 pKa = 9.39SKK515 pKa = 9.3QMRR518 pKa = 11.84GAMTGIDD525 pKa = 3.9NPYY528 pKa = 9.76VIQSATKK535 pKa = 8.27MM536 pKa = 3.57
MM1 pKa = 6.87NQALVILLVSFQLGVALDD19 pKa = 3.93NSVLAPIGVASAQEE33 pKa = 4.0WQLAAYY39 pKa = 6.49TTTLTGTIAVRR50 pKa = 11.84FIPVLPGNLSTCAQEE65 pKa = 4.48TLQEE69 pKa = 4.11YY70 pKa = 10.47NRR72 pKa = 11.84TVTNILGPLRR82 pKa = 11.84EE83 pKa = 4.36NLDD86 pKa = 3.68ALLSDD91 pKa = 4.23FDD93 pKa = 4.25KK94 pKa = 10.78PASRR98 pKa = 11.84FVGAIIGSVALGVATAAQITAAVALNQAQEE128 pKa = 3.82NARR131 pKa = 11.84NIWRR135 pKa = 11.84LKK137 pKa = 10.27EE138 pKa = 4.31SIKK141 pKa = 9.74KK142 pKa = 8.26TNEE145 pKa = 3.36AVLEE149 pKa = 4.42LKK151 pKa = 10.82DD152 pKa = 4.01GLATTAIALDD162 pKa = 3.89KK163 pKa = 10.27VQKK166 pKa = 10.5FINDD170 pKa = 4.08DD171 pKa = 4.22IIPQIKK177 pKa = 10.48DD178 pKa = 2.7IDD180 pKa = 4.17CQVVANKK187 pKa = 10.52LGVYY191 pKa = 10.44LSLYY195 pKa = 8.74LTEE198 pKa = 4.3LTTVFGSQITNPALSTLSYY217 pKa = 9.72QALYY221 pKa = 9.92SLCGGDD227 pKa = 3.27MGKK230 pKa = 8.33LTEE233 pKa = 5.08LIGVNAKK240 pKa = 10.31DD241 pKa = 3.4VGSLYY246 pKa = 10.33EE247 pKa = 4.26ANLITGQIVGYY258 pKa = 10.32DD259 pKa = 3.66PEE261 pKa = 4.29LQIILIQVSYY271 pKa = 10.28PSVSEE276 pKa = 4.1VTGVRR281 pKa = 11.84ATEE284 pKa = 4.01LVTVSVTTPKK294 pKa = 11.1GEE296 pKa = 4.01GQAIVPRR303 pKa = 11.84YY304 pKa = 7.88VAQSRR309 pKa = 11.84VLTEE313 pKa = 3.93EE314 pKa = 4.94LDD316 pKa = 3.61VSTCRR321 pKa = 11.84FSKK324 pKa = 7.66TTLYY328 pKa = 10.42CRR330 pKa = 11.84SILTRR335 pKa = 11.84PLPTLIASCLSGKK348 pKa = 10.3YY349 pKa = 9.82DD350 pKa = 3.52DD351 pKa = 4.82CQYY354 pKa = 8.24TTEE357 pKa = 4.23IGALSSRR364 pKa = 11.84FITVNGGVLANCRR377 pKa = 11.84AIVCKK382 pKa = 10.23CVSPPHH388 pKa = 7.16IIPQNDD394 pKa = 3.17IGSVTVIDD402 pKa = 3.88SSICKK407 pKa = 9.49EE408 pKa = 4.06VVLEE412 pKa = 4.16SVQLRR417 pKa = 11.84LEE419 pKa = 4.66GKK421 pKa = 10.18LSSQYY426 pKa = 10.48FSNVTIDD433 pKa = 3.86LSQITTSGSLDD444 pKa = 3.1ISSEE448 pKa = 3.63IGSINNTVNRR458 pKa = 11.84VDD460 pKa = 3.94EE461 pKa = 4.94LIKK464 pKa = 10.72EE465 pKa = 4.33SNEE468 pKa = 3.23WLNAVNPRR476 pKa = 11.84LVNNTSIIVLCVLAALIIVWLIALTVCFCYY506 pKa = 10.1SARR509 pKa = 11.84YY510 pKa = 8.12SAKK513 pKa = 9.39SKK515 pKa = 9.3QMRR518 pKa = 11.84GAMTGIDD525 pKa = 3.9NPYY528 pKa = 9.76VIQSATKK535 pKa = 8.27MM536 pKa = 3.57
Molecular weight: 57.75 kDa
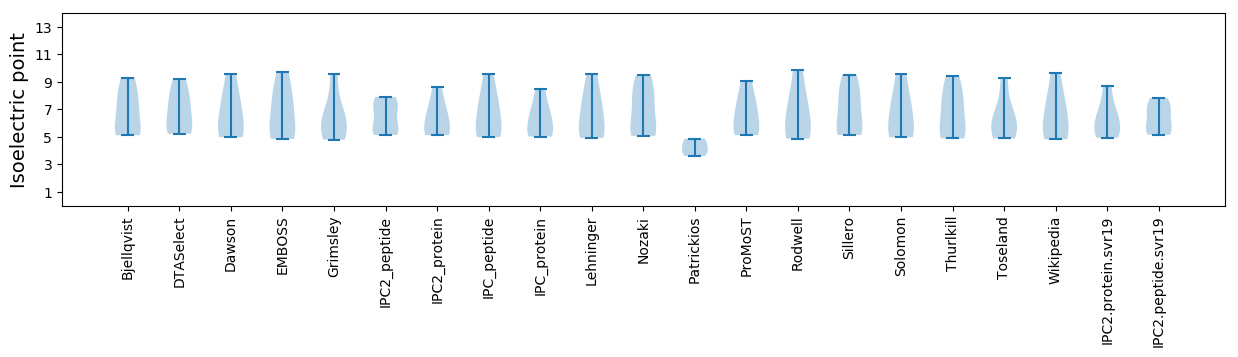
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F5BH21|F5BH21_9MONO Nucleocapsid OS=Avian metaavulavirus 2 OX=2560313 GN=NP PE=3 SV=1
MM1 pKa = 7.29AQTTVRR7 pKa = 11.84LYY9 pKa = 10.34IDD11 pKa = 3.69EE12 pKa = 4.89ASPDD16 pKa = 3.84IEE18 pKa = 4.26LLSYY22 pKa = 9.69PLIMKK27 pKa = 7.92DD28 pKa = 3.01TGHH31 pKa = 5.99GTKK34 pKa = 10.07EE35 pKa = 3.77LQQQIRR41 pKa = 11.84VAEE44 pKa = 4.21IGALQGGKK52 pKa = 9.74NEE54 pKa = 4.47SVFINAYY61 pKa = 10.42GFVQQCKK68 pKa = 8.58VKK70 pKa = 10.53PGATQFFQVDD80 pKa = 3.38AATKK84 pKa = 9.76PEE86 pKa = 4.3VVTAGMIIIGAVKK99 pKa = 9.95GVAGITKK106 pKa = 9.77LAEE109 pKa = 4.0EE110 pKa = 4.45VFEE113 pKa = 5.03LDD115 pKa = 3.82ISIKK119 pKa = 10.38KK120 pKa = 9.64SASFHH125 pKa = 5.93EE126 pKa = 4.62KK127 pKa = 10.09VAVSFNTVPLSLMNSTACRR146 pKa = 11.84NLGYY150 pKa = 8.83VTNAEE155 pKa = 4.15EE156 pKa = 5.34AIKK159 pKa = 10.62CPSKK163 pKa = 10.24IQAGVTYY170 pKa = 9.58KK171 pKa = 10.41FKK173 pKa = 11.01IMFVSLTRR181 pKa = 11.84LHH183 pKa = 6.25NGKK186 pKa = 10.26LYY188 pKa = 10.2RR189 pKa = 11.84VPKK192 pKa = 10.19AVYY195 pKa = 9.61AVEE198 pKa = 4.78ASALYY203 pKa = 9.93KK204 pKa = 10.58VQLEE208 pKa = 4.43VGFKK212 pKa = 10.63LDD214 pKa = 3.26VAKK217 pKa = 10.71DD218 pKa = 3.66HH219 pKa = 6.72PHH221 pKa = 6.0VKK223 pKa = 9.16MLKK226 pKa = 8.55KK227 pKa = 10.33VEE229 pKa = 4.37RR230 pKa = 11.84NGEE233 pKa = 4.16TLYY236 pKa = 11.14LGYY239 pKa = 11.09AWFHH243 pKa = 6.03LCNFKK248 pKa = 9.24KK249 pKa = 9.31TNAKK253 pKa = 10.21GEE255 pKa = 4.16SRR257 pKa = 11.84TISNLEE263 pKa = 3.82GKK265 pKa = 9.05VRR267 pKa = 11.84AMGIKK272 pKa = 10.16VSLYY276 pKa = 10.44DD277 pKa = 3.23LWGPTLVVQITGKK290 pKa = 7.51TSKK293 pKa = 10.03YY294 pKa = 9.15AQGFFSTTGTCCLPVSKK311 pKa = 10.42AAPEE315 pKa = 3.93LAKK318 pKa = 11.1LMWSCNATIVEE329 pKa = 4.28AAVIIQGSDD338 pKa = 2.52RR339 pKa = 11.84RR340 pKa = 11.84AVVTSEE346 pKa = 3.67DD347 pKa = 3.89LEE349 pKa = 4.82VYY351 pKa = 10.41GAVAKK356 pKa = 9.89EE357 pKa = 3.67KK358 pKa = 9.91QAAKK362 pKa = 10.18GFHH365 pKa = 7.1PFRR368 pKa = 11.84KK369 pKa = 9.78
MM1 pKa = 7.29AQTTVRR7 pKa = 11.84LYY9 pKa = 10.34IDD11 pKa = 3.69EE12 pKa = 4.89ASPDD16 pKa = 3.84IEE18 pKa = 4.26LLSYY22 pKa = 9.69PLIMKK27 pKa = 7.92DD28 pKa = 3.01TGHH31 pKa = 5.99GTKK34 pKa = 10.07EE35 pKa = 3.77LQQQIRR41 pKa = 11.84VAEE44 pKa = 4.21IGALQGGKK52 pKa = 9.74NEE54 pKa = 4.47SVFINAYY61 pKa = 10.42GFVQQCKK68 pKa = 8.58VKK70 pKa = 10.53PGATQFFQVDD80 pKa = 3.38AATKK84 pKa = 9.76PEE86 pKa = 4.3VVTAGMIIIGAVKK99 pKa = 9.95GVAGITKK106 pKa = 9.77LAEE109 pKa = 4.0EE110 pKa = 4.45VFEE113 pKa = 5.03LDD115 pKa = 3.82ISIKK119 pKa = 10.38KK120 pKa = 9.64SASFHH125 pKa = 5.93EE126 pKa = 4.62KK127 pKa = 10.09VAVSFNTVPLSLMNSTACRR146 pKa = 11.84NLGYY150 pKa = 8.83VTNAEE155 pKa = 4.15EE156 pKa = 5.34AIKK159 pKa = 10.62CPSKK163 pKa = 10.24IQAGVTYY170 pKa = 9.58KK171 pKa = 10.41FKK173 pKa = 11.01IMFVSLTRR181 pKa = 11.84LHH183 pKa = 6.25NGKK186 pKa = 10.26LYY188 pKa = 10.2RR189 pKa = 11.84VPKK192 pKa = 10.19AVYY195 pKa = 9.61AVEE198 pKa = 4.78ASALYY203 pKa = 9.93KK204 pKa = 10.58VQLEE208 pKa = 4.43VGFKK212 pKa = 10.63LDD214 pKa = 3.26VAKK217 pKa = 10.71DD218 pKa = 3.66HH219 pKa = 6.72PHH221 pKa = 6.0VKK223 pKa = 9.16MLKK226 pKa = 8.55KK227 pKa = 10.33VEE229 pKa = 4.37RR230 pKa = 11.84NGEE233 pKa = 4.16TLYY236 pKa = 11.14LGYY239 pKa = 11.09AWFHH243 pKa = 6.03LCNFKK248 pKa = 9.24KK249 pKa = 9.31TNAKK253 pKa = 10.21GEE255 pKa = 4.16SRR257 pKa = 11.84TISNLEE263 pKa = 3.82GKK265 pKa = 9.05VRR267 pKa = 11.84AMGIKK272 pKa = 10.16VSLYY276 pKa = 10.44DD277 pKa = 3.23LWGPTLVVQITGKK290 pKa = 7.51TSKK293 pKa = 10.03YY294 pKa = 9.15AQGFFSTTGTCCLPVSKK311 pKa = 10.42AAPEE315 pKa = 3.93LAKK318 pKa = 11.1LMWSCNATIVEE329 pKa = 4.28AAVIIQGSDD338 pKa = 2.52RR339 pKa = 11.84RR340 pKa = 11.84AVVTSEE346 pKa = 3.67DD347 pKa = 3.89LEE349 pKa = 4.82VYY351 pKa = 10.41GAVAKK356 pKa = 9.89EE357 pKa = 3.67KK358 pKa = 9.91QAAKK362 pKa = 10.18GFHH365 pKa = 7.1PFRR368 pKa = 11.84KK369 pKa = 9.78
Molecular weight: 40.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4583 |
369 |
2242 |
763.8 |
84.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.008 ± 0.937 | 1.898 ± 0.271 |
5.062 ± 0.419 | 5.062 ± 0.374 |
3.491 ± 0.461 | 5.651 ± 0.646 |
1.898 ± 0.412 | 7.004 ± 0.679 |
4.931 ± 0.792 | 10.168 ± 0.827 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.335 ± 0.378 | 4.32 ± 0.408 |
4.08 ± 0.385 | 4.757 ± 0.277 |
5.04 ± 0.511 | 8.662 ± 0.463 |
6.655 ± 0.479 | 6.917 ± 0.626 |
1.069 ± 0.131 | 2.989 ± 0.268 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
