
Neptunomonas qingdaonensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Oceanospirillaceae; Neptunomonas
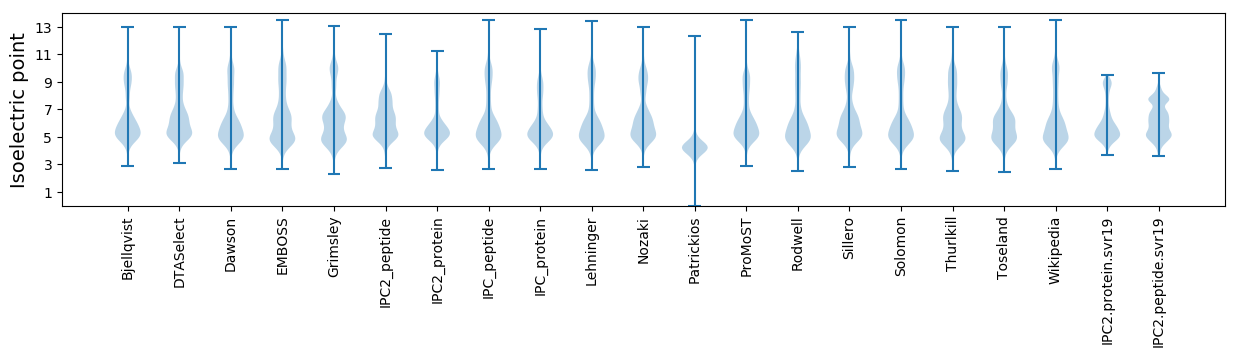
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
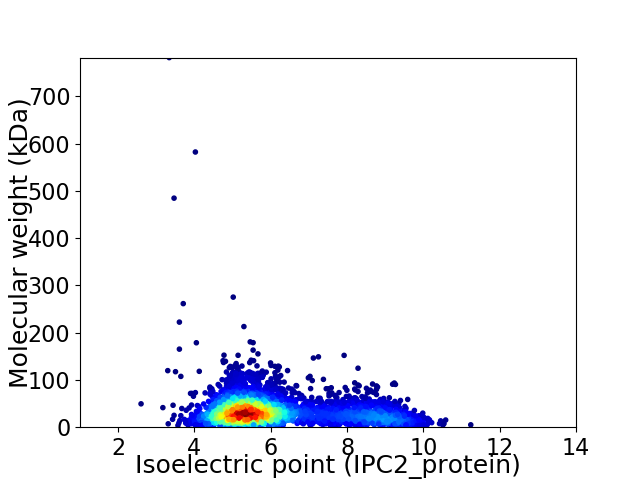
Virtual 2D-PAGE plot for 4011 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2LMB4|A0A1I2LMB4_9GAMM Simple sugar transport system substrate-binding protein OS=Neptunomonas qingdaonensis OX=1045558 GN=SAMN05216175_10138 PE=3 SV=1
NN1 pKa = 7.77GIQDD5 pKa = 3.53TGEE8 pKa = 4.26GGVAGAKK15 pKa = 9.95VSLFNCDD22 pKa = 2.22GTAANDD28 pKa = 3.48INGNPVASITTGGDD42 pKa = 3.27GIYY45 pKa = 10.72NFTNLAPGEE54 pKa = 4.35YY55 pKa = 8.92YY56 pKa = 10.98VQFTAPSGYY65 pKa = 10.88VITQQLATEE74 pKa = 4.6ADD76 pKa = 3.55AAGDD80 pKa = 4.0DD81 pKa = 4.1SDD83 pKa = 5.71ANAAGITSCVVLEE96 pKa = 4.26SGEE99 pKa = 4.17TEE101 pKa = 3.98EE102 pKa = 5.73RR103 pKa = 11.84VDD105 pKa = 5.0AGLVLQNAGIDD116 pKa = 3.19IEE118 pKa = 4.63KK119 pKa = 10.01YY120 pKa = 9.35VRR122 pKa = 11.84GRR124 pKa = 11.84YY125 pKa = 8.81EE126 pKa = 4.17GEE128 pKa = 4.02SGTDD132 pKa = 3.05EE133 pKa = 4.49GLTPGFWKK141 pKa = 8.45THH143 pKa = 5.13SEE145 pKa = 4.16FGPAPLAGWPEE156 pKa = 4.35SGFDD160 pKa = 3.48PTDD163 pKa = 3.34SYY165 pKa = 12.1EE166 pKa = 4.78SIFSVEE172 pKa = 4.28VPGTPSLLDD181 pKa = 3.66ALNSGGGDD189 pKa = 3.29LDD191 pKa = 3.43ALMRR195 pKa = 11.84HH196 pKa = 5.28STAALLNASNPFVAYY211 pKa = 9.2EE212 pKa = 3.73YY213 pKa = 10.77SQAEE217 pKa = 4.88VISMTQAAISSGVYY231 pKa = 10.21EE232 pKa = 3.89ITKK235 pKa = 10.62DD236 pKa = 3.18LFAAKK241 pKa = 10.26NEE243 pKa = 4.18LGADD247 pKa = 3.79LNEE250 pKa = 4.5GTSEE254 pKa = 4.03GTIVEE259 pKa = 4.73TIDD262 pKa = 3.57YY263 pKa = 10.68DD264 pKa = 4.26ADD266 pKa = 4.13TPDD269 pKa = 3.88GPYY272 pKa = 10.13IPIDD276 pKa = 3.45GEE278 pKa = 4.5AIFTYY283 pKa = 10.63VVTNTGDD290 pKa = 3.22VALSDD295 pKa = 3.78VQVTDD300 pKa = 4.13DD301 pKa = 3.47RR302 pKa = 11.84LANVTLVGGDD312 pKa = 3.68TNNDD316 pKa = 2.95NLLDD320 pKa = 3.92VGEE323 pKa = 4.38TWTYY327 pKa = 7.95TASEE331 pKa = 4.37TVVAGGTFVNTGTVTGADD349 pKa = 3.01ATGRR353 pKa = 11.84ILSDD357 pKa = 3.4SDD359 pKa = 3.55LAHH362 pKa = 6.67YY363 pKa = 7.51NTSFLTSSLGDD374 pKa = 3.78RR375 pKa = 11.84VWLDD379 pKa = 3.56LDD381 pKa = 3.91SDD383 pKa = 4.93GIQDD387 pKa = 3.48NDD389 pKa = 3.32EE390 pKa = 4.53AGVAGVTVNLLDD402 pKa = 3.69SDD404 pKa = 4.66GNVSATTITDD414 pKa = 3.26DD415 pKa = 3.37YY416 pKa = 11.58GNYY419 pKa = 10.16LFVALEE425 pKa = 4.14PGEE428 pKa = 4.12YY429 pKa = 9.69AIQVVAPDD437 pKa = 3.68GTVFTSQNQGMDD449 pKa = 3.79DD450 pKa = 4.51DD451 pKa = 5.18RR452 pKa = 11.84DD453 pKa = 3.67SDD455 pKa = 4.0VASNGVSEE463 pKa = 4.6FTTLTAGEE471 pKa = 4.86NDD473 pKa = 4.01LSWDD477 pKa = 3.44AGLVPTGTIDD487 pKa = 2.85IEE489 pKa = 4.27KK490 pKa = 9.71FVRR493 pKa = 11.84IEE495 pKa = 3.97TPGDD499 pKa = 3.51FFEE502 pKa = 5.88GAVCDD507 pKa = 4.05EE508 pKa = 4.57FGKK511 pKa = 9.37ATALTFDD518 pKa = 4.27YY519 pKa = 11.12EE520 pKa = 4.13EE521 pKa = 4.31STNVSTSQDD530 pKa = 3.05SSKK533 pKa = 11.53AKK535 pKa = 10.17ILTGTTFDD543 pKa = 4.51GDD545 pKa = 3.54GKK547 pKa = 10.99SYY549 pKa = 11.27VIVTDD554 pKa = 3.93KK555 pKa = 11.31SDD557 pKa = 3.42PTDD560 pKa = 3.64LGGKK564 pKa = 9.39VYY566 pKa = 10.45FSGYY570 pKa = 8.07VVEE573 pKa = 5.39GEE575 pKa = 4.43EE576 pKa = 4.34FTATAADD583 pKa = 3.55AGADD587 pKa = 3.31RR588 pKa = 11.84FGSNTFVHH596 pKa = 7.02FFDD599 pKa = 6.39DD600 pKa = 3.97NPFDD604 pKa = 4.16NVVGDD609 pKa = 4.69DD610 pKa = 5.7DD611 pKa = 5.46LLQSLTYY618 pKa = 8.87HH619 pKa = 6.46TSCSQPIYY627 pKa = 10.76LGDD630 pKa = 3.46QVGNVTLVGYY640 pKa = 10.02SGEE643 pKa = 4.35SGTAPSSPPVIGPDD657 pKa = 3.48LDD659 pKa = 4.96ADD661 pKa = 4.75TIAEE665 pKa = 4.41GPIGTIGTDD674 pKa = 3.29TAVFTYY680 pKa = 10.07KK681 pKa = 10.5VKK683 pKa = 9.67NTGTSDD689 pKa = 3.66LEE691 pKa = 4.37NVLVTDD697 pKa = 4.61DD698 pKa = 4.41RR699 pKa = 11.84LSEE702 pKa = 3.89LTLINKK708 pKa = 9.62GDD710 pKa = 3.91GDD712 pKa = 3.86NLLEE716 pKa = 4.9VGEE719 pKa = 4.32TWIYY723 pKa = 8.19TASEE727 pKa = 4.01AVLGGINTNIGKK739 pKa = 8.31VTANVVGTQFGVNDD753 pKa = 3.55QDD755 pKa = 3.31AANYY759 pKa = 10.3VGFDD763 pKa = 3.7TPPSIDD769 pKa = 3.19LEE771 pKa = 4.56KK772 pKa = 9.58YY773 pKa = 10.21VKK775 pKa = 10.79VEE777 pKa = 4.11GYY779 pKa = 8.57TPFVPMVCEE788 pKa = 4.27TYY790 pKa = 10.66GKK792 pKa = 8.53PVEE795 pKa = 4.06MTFVYY800 pKa = 10.33EE801 pKa = 3.99VDD803 pKa = 4.56DD804 pKa = 4.87DD805 pKa = 4.55NEE807 pKa = 4.98SISNTQEE814 pKa = 3.4GKK816 pKa = 10.79AKK818 pKa = 10.26ILTGTTLDD826 pKa = 3.86NDD828 pKa = 3.46DD829 pKa = 4.45LAWVRR834 pKa = 11.84VTDD837 pKa = 3.53SDD839 pKa = 4.03KK840 pKa = 11.62AFDD843 pKa = 4.19GGGKK847 pKa = 9.27IYY849 pKa = 10.8FEE851 pKa = 4.18GLVAKK856 pKa = 10.35GADD859 pKa = 3.83FTATAALAGADD870 pKa = 3.42RR871 pKa = 11.84FASSTYY877 pKa = 9.54IHH879 pKa = 6.56YY880 pKa = 10.61FDD882 pKa = 6.11DD883 pKa = 4.16EE884 pKa = 5.66ADD886 pKa = 3.62FTTEE890 pKa = 3.55NSGGLLQSLTYY901 pKa = 8.9HH902 pKa = 6.35TSCSKK907 pKa = 10.57PINLGDD913 pKa = 3.38EE914 pKa = 3.88TGYY917 pKa = 11.38AKK919 pKa = 10.73LVGYY923 pKa = 10.21VGEE926 pKa = 5.26DD927 pKa = 3.45GAAEE931 pKa = 3.89PVNEE935 pKa = 4.21YY936 pKa = 11.28GEE938 pKa = 4.79DD939 pKa = 3.69ADD941 pKa = 4.59EE942 pKa = 5.2GPGPEE947 pKa = 3.95AAFGSTVEE955 pKa = 4.05FTYY958 pKa = 11.06VVTNTGSTALGQIEE972 pKa = 4.85LSDD975 pKa = 3.97DD976 pKa = 4.29RR977 pKa = 11.84ISGLTLVGGDD987 pKa = 4.22DD988 pKa = 5.53DD989 pKa = 5.81GDD991 pKa = 3.75NLLDD995 pKa = 4.91VGEE998 pKa = 4.33EE999 pKa = 3.98WLFTGSEE1006 pKa = 4.33TAVSGQVTNIGKK1018 pKa = 8.44VTGTAVDD1025 pKa = 3.55EE1026 pKa = 4.93KK1027 pKa = 11.16GNSLNLPTVTDD1038 pKa = 4.05SDD1040 pKa = 3.72PGNYY1044 pKa = 9.07VVVGGPSIDD1053 pKa = 3.29VEE1055 pKa = 4.49KK1056 pKa = 10.55YY1057 pKa = 9.61VKK1059 pKa = 10.74VEE1061 pKa = 4.04GGGVPYY1067 pKa = 10.28EE1068 pKa = 4.15ALVCDD1073 pKa = 4.5TYY1075 pKa = 11.86GKK1077 pKa = 10.33ALAMTFRR1084 pKa = 11.84YY1085 pKa = 10.28LEE1087 pKa = 4.43GSTVDD1092 pKa = 3.29TDD1094 pKa = 3.42QDD1096 pKa = 3.57SGKK1099 pKa = 10.88AYY1101 pKa = 9.52IDD1103 pKa = 3.91KK1104 pKa = 11.01DD1105 pKa = 4.12LGTDD1109 pKa = 3.61DD1110 pKa = 6.02DD1111 pKa = 4.56GVSYY1115 pKa = 10.69IIVSDD1120 pKa = 3.6KK1121 pKa = 11.21SSPTDD1126 pKa = 3.41LSGKK1130 pKa = 10.06VYY1132 pKa = 10.34FQGEE1136 pKa = 4.39VAKK1139 pKa = 10.78GADD1142 pKa = 3.94FTALGASSFSSSTYY1156 pKa = 9.35IHH1158 pKa = 6.58YY1159 pKa = 10.64FDD1161 pKa = 6.1DD1162 pKa = 4.33SPFDD1166 pKa = 3.82GEE1168 pKa = 4.48GSIDD1172 pKa = 3.87LLQSSAYY1179 pKa = 8.26HH1180 pKa = 6.0TSCSQPINIGDD1191 pKa = 3.77VVGNTTLVGYY1201 pKa = 10.27LGEE1204 pKa = 5.09DD1205 pKa = 3.91GEE1207 pKa = 4.79ANPTGSNDD1215 pKa = 3.88GLGDD1219 pKa = 4.48DD1220 pKa = 5.35ADD1222 pKa = 4.25TGPGPDD1228 pKa = 3.44AAFGSDD1234 pKa = 3.27VVFNYY1239 pKa = 10.03VVTNTGSGALKK1250 pKa = 10.68NVVLTDD1256 pKa = 3.69DD1257 pKa = 5.06RR1258 pKa = 11.84ISDD1261 pKa = 3.6VTYY1264 pKa = 11.07VSGDD1268 pKa = 3.71DD1269 pKa = 3.66GDD1271 pKa = 4.27KK1272 pKa = 10.82LLEE1275 pKa = 4.43VGEE1278 pKa = 4.26EE1279 pKa = 3.8WLYY1282 pKa = 10.27TATEE1286 pKa = 4.06KK1287 pKa = 10.86AGQGVIGNIGKK1298 pKa = 8.04VTAKK1302 pKa = 10.45DD1303 pKa = 3.25AATGTISVMDD1313 pKa = 4.32KK1314 pKa = 10.98DD1315 pKa = 3.46AAFYY1319 pKa = 10.75NGSAPGITLGDD1330 pKa = 3.26RR1331 pKa = 11.84VWFDD1335 pKa = 3.83KK1336 pKa = 11.42NCDD1339 pKa = 3.67GIQDD1343 pKa = 4.03DD1344 pKa = 5.14GEE1346 pKa = 4.85NGAAGITVNLMNSVGNVIATTTTDD1370 pKa = 2.49VDD1372 pKa = 3.71GNYY1375 pKa = 10.36LFTDD1379 pKa = 4.73LAPGTYY1385 pKa = 9.85SVGFVAPNGYY1395 pKa = 9.66IFTKK1399 pKa = 10.44QDD1401 pKa = 2.76ATADD1405 pKa = 4.01DD1406 pKa = 4.11VDD1408 pKa = 4.49SDD1410 pKa = 4.26ANANGLTGQYY1420 pKa = 10.37KK1421 pKa = 10.33LAGGEE1426 pKa = 4.23TNLTVDD1432 pKa = 3.28AGLVEE1437 pKa = 4.8FTTKK1441 pKa = 10.54DD1442 pKa = 3.17VSFSFNGSSSGSGTYY1457 pKa = 11.04GNIRR1461 pKa = 11.84TYY1463 pKa = 10.36TDD1465 pKa = 2.7NGVSVNVSAHH1475 pKa = 5.22SRR1477 pKa = 11.84SDD1479 pKa = 3.56KK1480 pKa = 11.22SGDD1483 pKa = 2.86WSTSYY1488 pKa = 11.07LGAFSGGMGVTNRR1501 pKa = 11.84NEE1503 pKa = 4.17SGSGNSHH1510 pKa = 5.37TVDD1513 pKa = 3.24NRR1515 pKa = 11.84GDD1517 pKa = 3.29MDD1519 pKa = 5.16DD1520 pKa = 3.79YY1521 pKa = 11.92VLFEE1525 pKa = 4.13FDD1527 pKa = 3.47QNVVIDD1533 pKa = 4.13SAFLGYY1539 pKa = 10.6VVDD1542 pKa = 5.0DD1543 pKa = 3.91SDD1545 pKa = 3.6MSVWIGTLDD1554 pKa = 3.41NAFNNHH1560 pKa = 5.4TNLSDD1565 pKa = 5.94DD1566 pKa = 4.41ILSQMGFTEE1575 pKa = 4.34INN1577 pKa = 3.09
NN1 pKa = 7.77GIQDD5 pKa = 3.53TGEE8 pKa = 4.26GGVAGAKK15 pKa = 9.95VSLFNCDD22 pKa = 2.22GTAANDD28 pKa = 3.48INGNPVASITTGGDD42 pKa = 3.27GIYY45 pKa = 10.72NFTNLAPGEE54 pKa = 4.35YY55 pKa = 8.92YY56 pKa = 10.98VQFTAPSGYY65 pKa = 10.88VITQQLATEE74 pKa = 4.6ADD76 pKa = 3.55AAGDD80 pKa = 4.0DD81 pKa = 4.1SDD83 pKa = 5.71ANAAGITSCVVLEE96 pKa = 4.26SGEE99 pKa = 4.17TEE101 pKa = 3.98EE102 pKa = 5.73RR103 pKa = 11.84VDD105 pKa = 5.0AGLVLQNAGIDD116 pKa = 3.19IEE118 pKa = 4.63KK119 pKa = 10.01YY120 pKa = 9.35VRR122 pKa = 11.84GRR124 pKa = 11.84YY125 pKa = 8.81EE126 pKa = 4.17GEE128 pKa = 4.02SGTDD132 pKa = 3.05EE133 pKa = 4.49GLTPGFWKK141 pKa = 8.45THH143 pKa = 5.13SEE145 pKa = 4.16FGPAPLAGWPEE156 pKa = 4.35SGFDD160 pKa = 3.48PTDD163 pKa = 3.34SYY165 pKa = 12.1EE166 pKa = 4.78SIFSVEE172 pKa = 4.28VPGTPSLLDD181 pKa = 3.66ALNSGGGDD189 pKa = 3.29LDD191 pKa = 3.43ALMRR195 pKa = 11.84HH196 pKa = 5.28STAALLNASNPFVAYY211 pKa = 9.2EE212 pKa = 3.73YY213 pKa = 10.77SQAEE217 pKa = 4.88VISMTQAAISSGVYY231 pKa = 10.21EE232 pKa = 3.89ITKK235 pKa = 10.62DD236 pKa = 3.18LFAAKK241 pKa = 10.26NEE243 pKa = 4.18LGADD247 pKa = 3.79LNEE250 pKa = 4.5GTSEE254 pKa = 4.03GTIVEE259 pKa = 4.73TIDD262 pKa = 3.57YY263 pKa = 10.68DD264 pKa = 4.26ADD266 pKa = 4.13TPDD269 pKa = 3.88GPYY272 pKa = 10.13IPIDD276 pKa = 3.45GEE278 pKa = 4.5AIFTYY283 pKa = 10.63VVTNTGDD290 pKa = 3.22VALSDD295 pKa = 3.78VQVTDD300 pKa = 4.13DD301 pKa = 3.47RR302 pKa = 11.84LANVTLVGGDD312 pKa = 3.68TNNDD316 pKa = 2.95NLLDD320 pKa = 3.92VGEE323 pKa = 4.38TWTYY327 pKa = 7.95TASEE331 pKa = 4.37TVVAGGTFVNTGTVTGADD349 pKa = 3.01ATGRR353 pKa = 11.84ILSDD357 pKa = 3.4SDD359 pKa = 3.55LAHH362 pKa = 6.67YY363 pKa = 7.51NTSFLTSSLGDD374 pKa = 3.78RR375 pKa = 11.84VWLDD379 pKa = 3.56LDD381 pKa = 3.91SDD383 pKa = 4.93GIQDD387 pKa = 3.48NDD389 pKa = 3.32EE390 pKa = 4.53AGVAGVTVNLLDD402 pKa = 3.69SDD404 pKa = 4.66GNVSATTITDD414 pKa = 3.26DD415 pKa = 3.37YY416 pKa = 11.58GNYY419 pKa = 10.16LFVALEE425 pKa = 4.14PGEE428 pKa = 4.12YY429 pKa = 9.69AIQVVAPDD437 pKa = 3.68GTVFTSQNQGMDD449 pKa = 3.79DD450 pKa = 4.51DD451 pKa = 5.18RR452 pKa = 11.84DD453 pKa = 3.67SDD455 pKa = 4.0VASNGVSEE463 pKa = 4.6FTTLTAGEE471 pKa = 4.86NDD473 pKa = 4.01LSWDD477 pKa = 3.44AGLVPTGTIDD487 pKa = 2.85IEE489 pKa = 4.27KK490 pKa = 9.71FVRR493 pKa = 11.84IEE495 pKa = 3.97TPGDD499 pKa = 3.51FFEE502 pKa = 5.88GAVCDD507 pKa = 4.05EE508 pKa = 4.57FGKK511 pKa = 9.37ATALTFDD518 pKa = 4.27YY519 pKa = 11.12EE520 pKa = 4.13EE521 pKa = 4.31STNVSTSQDD530 pKa = 3.05SSKK533 pKa = 11.53AKK535 pKa = 10.17ILTGTTFDD543 pKa = 4.51GDD545 pKa = 3.54GKK547 pKa = 10.99SYY549 pKa = 11.27VIVTDD554 pKa = 3.93KK555 pKa = 11.31SDD557 pKa = 3.42PTDD560 pKa = 3.64LGGKK564 pKa = 9.39VYY566 pKa = 10.45FSGYY570 pKa = 8.07VVEE573 pKa = 5.39GEE575 pKa = 4.43EE576 pKa = 4.34FTATAADD583 pKa = 3.55AGADD587 pKa = 3.31RR588 pKa = 11.84FGSNTFVHH596 pKa = 7.02FFDD599 pKa = 6.39DD600 pKa = 3.97NPFDD604 pKa = 4.16NVVGDD609 pKa = 4.69DD610 pKa = 5.7DD611 pKa = 5.46LLQSLTYY618 pKa = 8.87HH619 pKa = 6.46TSCSQPIYY627 pKa = 10.76LGDD630 pKa = 3.46QVGNVTLVGYY640 pKa = 10.02SGEE643 pKa = 4.35SGTAPSSPPVIGPDD657 pKa = 3.48LDD659 pKa = 4.96ADD661 pKa = 4.75TIAEE665 pKa = 4.41GPIGTIGTDD674 pKa = 3.29TAVFTYY680 pKa = 10.07KK681 pKa = 10.5VKK683 pKa = 9.67NTGTSDD689 pKa = 3.66LEE691 pKa = 4.37NVLVTDD697 pKa = 4.61DD698 pKa = 4.41RR699 pKa = 11.84LSEE702 pKa = 3.89LTLINKK708 pKa = 9.62GDD710 pKa = 3.91GDD712 pKa = 3.86NLLEE716 pKa = 4.9VGEE719 pKa = 4.32TWIYY723 pKa = 8.19TASEE727 pKa = 4.01AVLGGINTNIGKK739 pKa = 8.31VTANVVGTQFGVNDD753 pKa = 3.55QDD755 pKa = 3.31AANYY759 pKa = 10.3VGFDD763 pKa = 3.7TPPSIDD769 pKa = 3.19LEE771 pKa = 4.56KK772 pKa = 9.58YY773 pKa = 10.21VKK775 pKa = 10.79VEE777 pKa = 4.11GYY779 pKa = 8.57TPFVPMVCEE788 pKa = 4.27TYY790 pKa = 10.66GKK792 pKa = 8.53PVEE795 pKa = 4.06MTFVYY800 pKa = 10.33EE801 pKa = 3.99VDD803 pKa = 4.56DD804 pKa = 4.87DD805 pKa = 4.55NEE807 pKa = 4.98SISNTQEE814 pKa = 3.4GKK816 pKa = 10.79AKK818 pKa = 10.26ILTGTTLDD826 pKa = 3.86NDD828 pKa = 3.46DD829 pKa = 4.45LAWVRR834 pKa = 11.84VTDD837 pKa = 3.53SDD839 pKa = 4.03KK840 pKa = 11.62AFDD843 pKa = 4.19GGGKK847 pKa = 9.27IYY849 pKa = 10.8FEE851 pKa = 4.18GLVAKK856 pKa = 10.35GADD859 pKa = 3.83FTATAALAGADD870 pKa = 3.42RR871 pKa = 11.84FASSTYY877 pKa = 9.54IHH879 pKa = 6.56YY880 pKa = 10.61FDD882 pKa = 6.11DD883 pKa = 4.16EE884 pKa = 5.66ADD886 pKa = 3.62FTTEE890 pKa = 3.55NSGGLLQSLTYY901 pKa = 8.9HH902 pKa = 6.35TSCSKK907 pKa = 10.57PINLGDD913 pKa = 3.38EE914 pKa = 3.88TGYY917 pKa = 11.38AKK919 pKa = 10.73LVGYY923 pKa = 10.21VGEE926 pKa = 5.26DD927 pKa = 3.45GAAEE931 pKa = 3.89PVNEE935 pKa = 4.21YY936 pKa = 11.28GEE938 pKa = 4.79DD939 pKa = 3.69ADD941 pKa = 4.59EE942 pKa = 5.2GPGPEE947 pKa = 3.95AAFGSTVEE955 pKa = 4.05FTYY958 pKa = 11.06VVTNTGSTALGQIEE972 pKa = 4.85LSDD975 pKa = 3.97DD976 pKa = 4.29RR977 pKa = 11.84ISGLTLVGGDD987 pKa = 4.22DD988 pKa = 5.53DD989 pKa = 5.81GDD991 pKa = 3.75NLLDD995 pKa = 4.91VGEE998 pKa = 4.33EE999 pKa = 3.98WLFTGSEE1006 pKa = 4.33TAVSGQVTNIGKK1018 pKa = 8.44VTGTAVDD1025 pKa = 3.55EE1026 pKa = 4.93KK1027 pKa = 11.16GNSLNLPTVTDD1038 pKa = 4.05SDD1040 pKa = 3.72PGNYY1044 pKa = 9.07VVVGGPSIDD1053 pKa = 3.29VEE1055 pKa = 4.49KK1056 pKa = 10.55YY1057 pKa = 9.61VKK1059 pKa = 10.74VEE1061 pKa = 4.04GGGVPYY1067 pKa = 10.28EE1068 pKa = 4.15ALVCDD1073 pKa = 4.5TYY1075 pKa = 11.86GKK1077 pKa = 10.33ALAMTFRR1084 pKa = 11.84YY1085 pKa = 10.28LEE1087 pKa = 4.43GSTVDD1092 pKa = 3.29TDD1094 pKa = 3.42QDD1096 pKa = 3.57SGKK1099 pKa = 10.88AYY1101 pKa = 9.52IDD1103 pKa = 3.91KK1104 pKa = 11.01DD1105 pKa = 4.12LGTDD1109 pKa = 3.61DD1110 pKa = 6.02DD1111 pKa = 4.56GVSYY1115 pKa = 10.69IIVSDD1120 pKa = 3.6KK1121 pKa = 11.21SSPTDD1126 pKa = 3.41LSGKK1130 pKa = 10.06VYY1132 pKa = 10.34FQGEE1136 pKa = 4.39VAKK1139 pKa = 10.78GADD1142 pKa = 3.94FTALGASSFSSSTYY1156 pKa = 9.35IHH1158 pKa = 6.58YY1159 pKa = 10.64FDD1161 pKa = 6.1DD1162 pKa = 4.33SPFDD1166 pKa = 3.82GEE1168 pKa = 4.48GSIDD1172 pKa = 3.87LLQSSAYY1179 pKa = 8.26HH1180 pKa = 6.0TSCSQPINIGDD1191 pKa = 3.77VVGNTTLVGYY1201 pKa = 10.27LGEE1204 pKa = 5.09DD1205 pKa = 3.91GEE1207 pKa = 4.79ANPTGSNDD1215 pKa = 3.88GLGDD1219 pKa = 4.48DD1220 pKa = 5.35ADD1222 pKa = 4.25TGPGPDD1228 pKa = 3.44AAFGSDD1234 pKa = 3.27VVFNYY1239 pKa = 10.03VVTNTGSGALKK1250 pKa = 10.68NVVLTDD1256 pKa = 3.69DD1257 pKa = 5.06RR1258 pKa = 11.84ISDD1261 pKa = 3.6VTYY1264 pKa = 11.07VSGDD1268 pKa = 3.71DD1269 pKa = 3.66GDD1271 pKa = 4.27KK1272 pKa = 10.82LLEE1275 pKa = 4.43VGEE1278 pKa = 4.26EE1279 pKa = 3.8WLYY1282 pKa = 10.27TATEE1286 pKa = 4.06KK1287 pKa = 10.86AGQGVIGNIGKK1298 pKa = 8.04VTAKK1302 pKa = 10.45DD1303 pKa = 3.25AATGTISVMDD1313 pKa = 4.32KK1314 pKa = 10.98DD1315 pKa = 3.46AAFYY1319 pKa = 10.75NGSAPGITLGDD1330 pKa = 3.26RR1331 pKa = 11.84VWFDD1335 pKa = 3.83KK1336 pKa = 11.42NCDD1339 pKa = 3.67GIQDD1343 pKa = 4.03DD1344 pKa = 5.14GEE1346 pKa = 4.85NGAAGITVNLMNSVGNVIATTTTDD1370 pKa = 2.49VDD1372 pKa = 3.71GNYY1375 pKa = 10.36LFTDD1379 pKa = 4.73LAPGTYY1385 pKa = 9.85SVGFVAPNGYY1395 pKa = 9.66IFTKK1399 pKa = 10.44QDD1401 pKa = 2.76ATADD1405 pKa = 4.01DD1406 pKa = 4.11VDD1408 pKa = 4.49SDD1410 pKa = 4.26ANANGLTGQYY1420 pKa = 10.37KK1421 pKa = 10.33LAGGEE1426 pKa = 4.23TNLTVDD1432 pKa = 3.28AGLVEE1437 pKa = 4.8FTTKK1441 pKa = 10.54DD1442 pKa = 3.17VSFSFNGSSSGSGTYY1457 pKa = 11.04GNIRR1461 pKa = 11.84TYY1463 pKa = 10.36TDD1465 pKa = 2.7NGVSVNVSAHH1475 pKa = 5.22SRR1477 pKa = 11.84SDD1479 pKa = 3.56KK1480 pKa = 11.22SGDD1483 pKa = 2.86WSTSYY1488 pKa = 11.07LGAFSGGMGVTNRR1501 pKa = 11.84NEE1503 pKa = 4.17SGSGNSHH1510 pKa = 5.37TVDD1513 pKa = 3.24NRR1515 pKa = 11.84GDD1517 pKa = 3.29MDD1519 pKa = 5.16DD1520 pKa = 3.79YY1521 pKa = 11.92VLFEE1525 pKa = 4.13FDD1527 pKa = 3.47QNVVIDD1533 pKa = 4.13SAFLGYY1539 pKa = 10.6VVDD1542 pKa = 5.0DD1543 pKa = 3.91SDD1545 pKa = 3.6MSVWIGTLDD1554 pKa = 3.41NAFNNHH1560 pKa = 5.4TNLSDD1565 pKa = 5.94DD1566 pKa = 4.41ILSQMGFTEE1575 pKa = 4.34INN1577 pKa = 3.09
Molecular weight: 165.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2LQ04|A0A1I2LQ04_9GAMM NADP-dependent 3-hydroxy acid dehydrogenase YdfG OS=Neptunomonas qingdaonensis OX=1045558 GN=SAMN05216175_101181 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.31SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.27GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.31SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.27GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1312960 |
24 |
7596 |
327.3 |
36.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.019 ± 0.049 | 1.027 ± 0.015 |
5.611 ± 0.052 | 6.08 ± 0.045 |
3.931 ± 0.025 | 7.001 ± 0.047 |
2.264 ± 0.022 | 6.43 ± 0.03 |
4.824 ± 0.042 | 10.735 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.028 | 3.922 ± 0.039 |
4.081 ± 0.027 | 4.477 ± 0.031 |
4.964 ± 0.042 | 6.714 ± 0.042 |
5.321 ± 0.063 | 6.928 ± 0.033 |
1.241 ± 0.016 | 2.833 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
