
Microbacterium sp. Gd 4-13
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
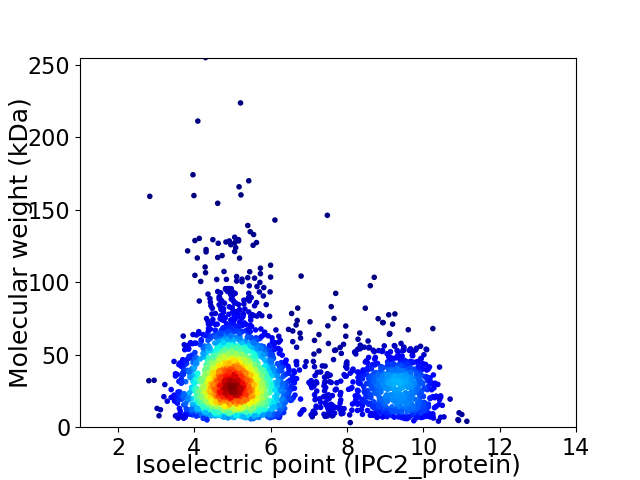
Virtual 2D-PAGE plot for 3162 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U0H0I4|A0A2U0H0I4_9MICO FMN-binding protein OS=Microbacterium sp. Gd 4-13 OX=2173179 GN=DEA06_15820 PE=4 SV=1
MM1 pKa = 6.86TRR3 pKa = 11.84VRR5 pKa = 11.84SRR7 pKa = 11.84IAVAVAVLTTVVLVASGCSVFTSDD31 pKa = 4.27SWAPPPSVTPDD42 pKa = 3.31VSGVSADD49 pKa = 4.13LLPFYY54 pKa = 10.63DD55 pKa = 5.25QDD57 pKa = 4.27VAWEE61 pKa = 4.23SCAAEE66 pKa = 4.02QQFDD70 pKa = 4.03CANVRR75 pKa = 11.84VPRR78 pKa = 11.84DD79 pKa = 3.1WSAPASGEE87 pKa = 3.69IEE89 pKa = 3.84IAVIRR94 pKa = 11.84HH95 pKa = 5.81RR96 pKa = 11.84ADD98 pKa = 2.73SGAPIGSLFTNPGGPGVSGVDD119 pKa = 3.44TLKK122 pKa = 10.7QALDD126 pKa = 3.8VVAGSPLRR134 pKa = 11.84EE135 pKa = 4.09NYY137 pKa = 10.19DD138 pKa = 3.32VIGFDD143 pKa = 3.56PRR145 pKa = 11.84GVGASTAIDD154 pKa = 4.24CYY156 pKa = 10.8DD157 pKa = 3.54TADD160 pKa = 4.17LDD162 pKa = 3.67QYY164 pKa = 11.54LYY166 pKa = 10.24DD167 pKa = 4.61IPDD170 pKa = 3.67APRR173 pKa = 11.84GSDD176 pKa = 2.58EE177 pKa = 3.7WTAEE181 pKa = 3.52IEE183 pKa = 4.19ARR185 pKa = 11.84DD186 pKa = 3.64AAFAAACDD194 pKa = 3.97ANSDD198 pKa = 3.66GLLPFVSTEE207 pKa = 3.47NAARR211 pKa = 11.84DD212 pKa = 3.92LDD214 pKa = 3.83VLRR217 pKa = 11.84AVMGEE222 pKa = 4.0TTLDD226 pKa = 3.37YY227 pKa = 11.17LGYY230 pKa = 10.35SWGTALGAAYY240 pKa = 10.32AEE242 pKa = 4.19LHH244 pKa = 6.65PDD246 pKa = 2.81RR247 pKa = 11.84VGRR250 pKa = 11.84MVLDD254 pKa = 4.26GALDD258 pKa = 3.74PSIPGAEE265 pKa = 3.97VGVGQMEE272 pKa = 4.91GFQRR276 pKa = 11.84SLDD279 pKa = 3.76AFLTACLSFDD289 pKa = 3.68DD290 pKa = 4.66CPFRR294 pKa = 11.84GTPAEE299 pKa = 4.06ASADD303 pKa = 3.71LDD305 pKa = 3.82ALFASVDD312 pKa = 3.29ARR314 pKa = 11.84PIVAADD320 pKa = 3.47GRR322 pKa = 11.84EE323 pKa = 4.03LGADD327 pKa = 3.42TLMLALLNALYY338 pKa = 10.66SPASWPYY345 pKa = 11.22LRR347 pKa = 11.84VTLSGVQNADD357 pKa = 3.47PAPAFTLADD366 pKa = 4.08AYY368 pKa = 10.77NARR371 pKa = 11.84QGGDD375 pKa = 3.22YY376 pKa = 10.83VGNSEE381 pKa = 4.61EE382 pKa = 5.03AFPAYY387 pKa = 11.02NCMDD391 pKa = 3.66YY392 pKa = 10.48PAVGEE397 pKa = 4.41AQTQAAQAEE406 pKa = 4.58LEE408 pKa = 4.32RR409 pKa = 11.84VAPLAAEE416 pKa = 4.35YY417 pKa = 10.57FSGPDD422 pKa = 3.09VCEE425 pKa = 3.67QWPYY429 pKa = 10.83PPTGTRR435 pKa = 11.84GPVSADD441 pKa = 2.7GAAPILVIGTTSDD454 pKa = 3.36PATPYY459 pKa = 9.9QWAVSLAEE467 pKa = 3.92QLTSGVLVTRR477 pKa = 11.84VGEE480 pKa = 4.08GHH482 pKa = 6.17TGYY485 pKa = 11.13NKK487 pKa = 10.76GNTCVDD493 pKa = 4.01DD494 pKa = 4.58AVVAYY499 pKa = 10.15LVDD502 pKa = 4.1GVVPDD507 pKa = 3.83ADD509 pKa = 3.63IRR511 pKa = 11.84CEE513 pKa = 3.76
MM1 pKa = 6.86TRR3 pKa = 11.84VRR5 pKa = 11.84SRR7 pKa = 11.84IAVAVAVLTTVVLVASGCSVFTSDD31 pKa = 4.27SWAPPPSVTPDD42 pKa = 3.31VSGVSADD49 pKa = 4.13LLPFYY54 pKa = 10.63DD55 pKa = 5.25QDD57 pKa = 4.27VAWEE61 pKa = 4.23SCAAEE66 pKa = 4.02QQFDD70 pKa = 4.03CANVRR75 pKa = 11.84VPRR78 pKa = 11.84DD79 pKa = 3.1WSAPASGEE87 pKa = 3.69IEE89 pKa = 3.84IAVIRR94 pKa = 11.84HH95 pKa = 5.81RR96 pKa = 11.84ADD98 pKa = 2.73SGAPIGSLFTNPGGPGVSGVDD119 pKa = 3.44TLKK122 pKa = 10.7QALDD126 pKa = 3.8VVAGSPLRR134 pKa = 11.84EE135 pKa = 4.09NYY137 pKa = 10.19DD138 pKa = 3.32VIGFDD143 pKa = 3.56PRR145 pKa = 11.84GVGASTAIDD154 pKa = 4.24CYY156 pKa = 10.8DD157 pKa = 3.54TADD160 pKa = 4.17LDD162 pKa = 3.67QYY164 pKa = 11.54LYY166 pKa = 10.24DD167 pKa = 4.61IPDD170 pKa = 3.67APRR173 pKa = 11.84GSDD176 pKa = 2.58EE177 pKa = 3.7WTAEE181 pKa = 3.52IEE183 pKa = 4.19ARR185 pKa = 11.84DD186 pKa = 3.64AAFAAACDD194 pKa = 3.97ANSDD198 pKa = 3.66GLLPFVSTEE207 pKa = 3.47NAARR211 pKa = 11.84DD212 pKa = 3.92LDD214 pKa = 3.83VLRR217 pKa = 11.84AVMGEE222 pKa = 4.0TTLDD226 pKa = 3.37YY227 pKa = 11.17LGYY230 pKa = 10.35SWGTALGAAYY240 pKa = 10.32AEE242 pKa = 4.19LHH244 pKa = 6.65PDD246 pKa = 2.81RR247 pKa = 11.84VGRR250 pKa = 11.84MVLDD254 pKa = 4.26GALDD258 pKa = 3.74PSIPGAEE265 pKa = 3.97VGVGQMEE272 pKa = 4.91GFQRR276 pKa = 11.84SLDD279 pKa = 3.76AFLTACLSFDD289 pKa = 3.68DD290 pKa = 4.66CPFRR294 pKa = 11.84GTPAEE299 pKa = 4.06ASADD303 pKa = 3.71LDD305 pKa = 3.82ALFASVDD312 pKa = 3.29ARR314 pKa = 11.84PIVAADD320 pKa = 3.47GRR322 pKa = 11.84EE323 pKa = 4.03LGADD327 pKa = 3.42TLMLALLNALYY338 pKa = 10.66SPASWPYY345 pKa = 11.22LRR347 pKa = 11.84VTLSGVQNADD357 pKa = 3.47PAPAFTLADD366 pKa = 4.08AYY368 pKa = 10.77NARR371 pKa = 11.84QGGDD375 pKa = 3.22YY376 pKa = 10.83VGNSEE381 pKa = 4.61EE382 pKa = 5.03AFPAYY387 pKa = 11.02NCMDD391 pKa = 3.66YY392 pKa = 10.48PAVGEE397 pKa = 4.41AQTQAAQAEE406 pKa = 4.58LEE408 pKa = 4.32RR409 pKa = 11.84VAPLAAEE416 pKa = 4.35YY417 pKa = 10.57FSGPDD422 pKa = 3.09VCEE425 pKa = 3.67QWPYY429 pKa = 10.83PPTGTRR435 pKa = 11.84GPVSADD441 pKa = 2.7GAAPILVIGTTSDD454 pKa = 3.36PATPYY459 pKa = 9.9QWAVSLAEE467 pKa = 3.92QLTSGVLVTRR477 pKa = 11.84VGEE480 pKa = 4.08GHH482 pKa = 6.17TGYY485 pKa = 11.13NKK487 pKa = 10.76GNTCVDD493 pKa = 4.01DD494 pKa = 4.58AVVAYY499 pKa = 10.15LVDD502 pKa = 4.1GVVPDD507 pKa = 3.83ADD509 pKa = 3.63IRR511 pKa = 11.84CEE513 pKa = 3.76
Molecular weight: 53.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U0H5U6|A0A2U0H5U6_9MICO Uncharacterized protein OS=Microbacterium sp. Gd 4-13 OX=2173179 GN=DEA06_09495 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1028731 |
29 |
2440 |
325.3 |
34.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.739 ± 0.058 | 0.471 ± 0.009 |
6.445 ± 0.042 | 5.547 ± 0.038 |
3.087 ± 0.027 | 8.935 ± 0.034 |
1.91 ± 0.021 | 4.452 ± 0.031 |
1.754 ± 0.031 | 9.87 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.719 ± 0.016 | 1.89 ± 0.026 |
5.45 ± 0.03 | 2.625 ± 0.023 |
7.535 ± 0.056 | 5.772 ± 0.028 |
6.13 ± 0.041 | 9.228 ± 0.044 |
1.513 ± 0.018 | 1.929 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
