
Alternanthera yellow vein virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.14
Get precalculated fractions of proteins
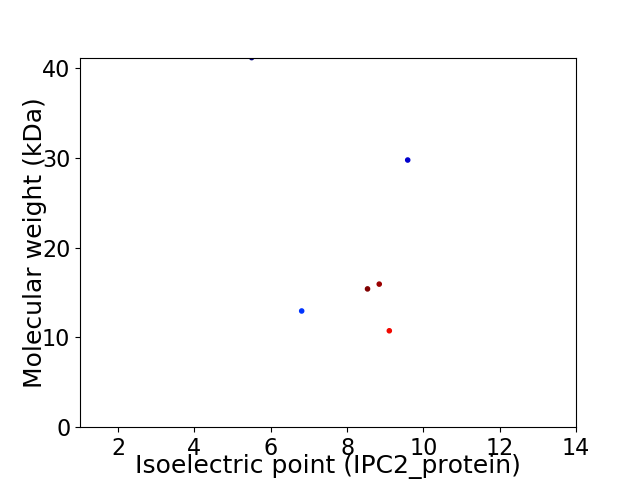
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
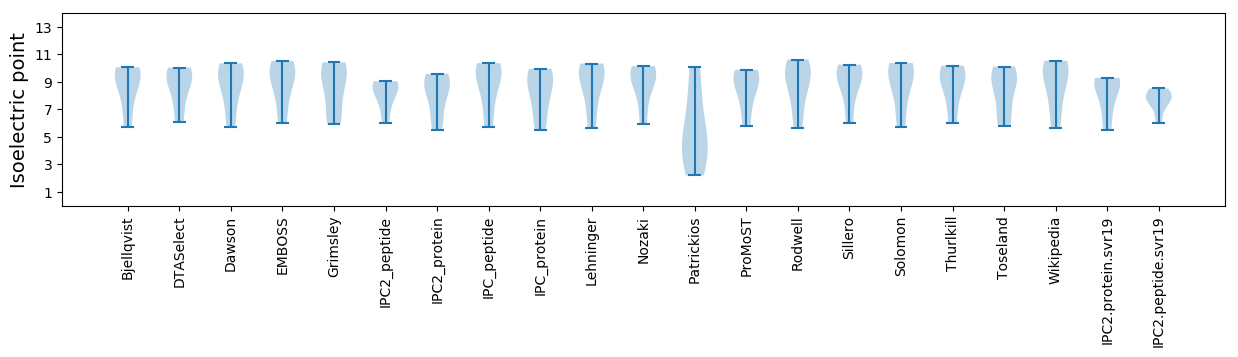
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4GZI6|Q4GZI6_9GEMI Transcriptional activator protein OS=Alternanthera yellow vein virus OX=337826 GN=ac2 PE=3 SV=1
MM1 pKa = 7.42RR2 pKa = 11.84QPGRR6 pKa = 11.84FRR8 pKa = 11.84LNRR11 pKa = 11.84KK12 pKa = 8.78NFFLTYY18 pKa = 8.51SQCPISKK25 pKa = 10.43EE26 pKa = 3.82EE27 pKa = 4.38ALEE30 pKa = 3.8QLLNIQTPVNKK41 pKa = 10.14KK42 pKa = 9.77FIRR45 pKa = 11.84VCRR48 pKa = 11.84EE49 pKa = 3.34LHH51 pKa = 6.55EE52 pKa = 5.57DD53 pKa = 3.97GNPHH57 pKa = 6.43LHH59 pKa = 6.93ALVQFEE65 pKa = 4.92GKK67 pKa = 10.14FDD69 pKa = 3.56LTNPRR74 pKa = 11.84FFDD77 pKa = 3.64LQSPNSSSQYY87 pKa = 10.25HH88 pKa = 6.03CFITDD93 pKa = 3.38AKK95 pKa = 10.73SSSDD99 pKa = 3.1VKK101 pKa = 11.12AYY103 pKa = 9.75IEE105 pKa = 4.26KK106 pKa = 10.91DD107 pKa = 3.08GDD109 pKa = 3.6ILDD112 pKa = 3.87WGTFQIDD119 pKa = 3.03GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 3.71AYY134 pKa = 10.81ARR136 pKa = 11.84AINTGNKK143 pKa = 9.39DD144 pKa = 3.15QALDD148 pKa = 3.97VIKK151 pKa = 10.51EE152 pKa = 4.1LAPKK156 pKa = 10.3DD157 pKa = 3.74YY158 pKa = 11.2VLQFHH163 pKa = 6.76NLNANLEE170 pKa = 4.56RR171 pKa = 11.84IFMPPIEE178 pKa = 4.63EE179 pKa = 4.31YY180 pKa = 10.23ISPFNCSSFDD190 pKa = 3.55QVPEE194 pKa = 4.52EE195 pKa = 4.12IEE197 pKa = 3.61EE198 pKa = 4.14WAADD202 pKa = 3.64NVMSAAARR210 pKa = 11.84PWRR213 pKa = 11.84PISIVIEE220 pKa = 4.23GDD222 pKa = 3.2SRR224 pKa = 11.84TGKK227 pKa = 8.52TMWARR232 pKa = 11.84SLGPHH237 pKa = 6.7NYY239 pKa = 10.01LCGHH243 pKa = 7.13IDD245 pKa = 3.96LSPRR249 pKa = 11.84VYY251 pKa = 11.1SNEE254 pKa = 3.04AWYY257 pKa = 10.37NIIDD261 pKa = 4.53DD262 pKa = 4.1VDD264 pKa = 3.43PHH266 pKa = 5.9YY267 pKa = 11.29VKK269 pKa = 10.59HH270 pKa = 6.1FKK272 pKa = 10.84EE273 pKa = 4.16FMGAQRR279 pKa = 11.84DD280 pKa = 3.81WQSNTKK286 pKa = 9.6YY287 pKa = 10.57GKK289 pKa = 9.12PVQIKK294 pKa = 10.41GGIPTIFLCNPGPTSSYY311 pKa = 11.31KK312 pKa = 10.67EE313 pKa = 3.89FLDD316 pKa = 3.57EE317 pKa = 4.57EE318 pKa = 4.61KK319 pKa = 11.2NIGLKK324 pKa = 9.82NWAIKK329 pKa = 10.38NATFVTINQPLYY341 pKa = 10.4SGSHH345 pKa = 4.95QSPTQASQEE354 pKa = 4.2EE355 pKa = 4.63TSQTEE360 pKa = 4.1GG361 pKa = 3.22
MM1 pKa = 7.42RR2 pKa = 11.84QPGRR6 pKa = 11.84FRR8 pKa = 11.84LNRR11 pKa = 11.84KK12 pKa = 8.78NFFLTYY18 pKa = 8.51SQCPISKK25 pKa = 10.43EE26 pKa = 3.82EE27 pKa = 4.38ALEE30 pKa = 3.8QLLNIQTPVNKK41 pKa = 10.14KK42 pKa = 9.77FIRR45 pKa = 11.84VCRR48 pKa = 11.84EE49 pKa = 3.34LHH51 pKa = 6.55EE52 pKa = 5.57DD53 pKa = 3.97GNPHH57 pKa = 6.43LHH59 pKa = 6.93ALVQFEE65 pKa = 4.92GKK67 pKa = 10.14FDD69 pKa = 3.56LTNPRR74 pKa = 11.84FFDD77 pKa = 3.64LQSPNSSSQYY87 pKa = 10.25HH88 pKa = 6.03CFITDD93 pKa = 3.38AKK95 pKa = 10.73SSSDD99 pKa = 3.1VKK101 pKa = 11.12AYY103 pKa = 9.75IEE105 pKa = 4.26KK106 pKa = 10.91DD107 pKa = 3.08GDD109 pKa = 3.6ILDD112 pKa = 3.87WGTFQIDD119 pKa = 3.03GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 3.71AYY134 pKa = 10.81ARR136 pKa = 11.84AINTGNKK143 pKa = 9.39DD144 pKa = 3.15QALDD148 pKa = 3.97VIKK151 pKa = 10.51EE152 pKa = 4.1LAPKK156 pKa = 10.3DD157 pKa = 3.74YY158 pKa = 11.2VLQFHH163 pKa = 6.76NLNANLEE170 pKa = 4.56RR171 pKa = 11.84IFMPPIEE178 pKa = 4.63EE179 pKa = 4.31YY180 pKa = 10.23ISPFNCSSFDD190 pKa = 3.55QVPEE194 pKa = 4.52EE195 pKa = 4.12IEE197 pKa = 3.61EE198 pKa = 4.14WAADD202 pKa = 3.64NVMSAAARR210 pKa = 11.84PWRR213 pKa = 11.84PISIVIEE220 pKa = 4.23GDD222 pKa = 3.2SRR224 pKa = 11.84TGKK227 pKa = 8.52TMWARR232 pKa = 11.84SLGPHH237 pKa = 6.7NYY239 pKa = 10.01LCGHH243 pKa = 7.13IDD245 pKa = 3.96LSPRR249 pKa = 11.84VYY251 pKa = 11.1SNEE254 pKa = 3.04AWYY257 pKa = 10.37NIIDD261 pKa = 4.53DD262 pKa = 4.1VDD264 pKa = 3.43PHH266 pKa = 5.9YY267 pKa = 11.29VKK269 pKa = 10.59HH270 pKa = 6.1FKK272 pKa = 10.84EE273 pKa = 4.16FMGAQRR279 pKa = 11.84DD280 pKa = 3.81WQSNTKK286 pKa = 9.6YY287 pKa = 10.57GKK289 pKa = 9.12PVQIKK294 pKa = 10.41GGIPTIFLCNPGPTSSYY311 pKa = 11.31KK312 pKa = 10.67EE313 pKa = 3.89FLDD316 pKa = 3.57EE317 pKa = 4.57EE318 pKa = 4.61KK319 pKa = 11.2NIGLKK324 pKa = 9.82NWAIKK329 pKa = 10.38NATFVTINQPLYY341 pKa = 10.4SGSHH345 pKa = 4.95QSPTQASQEE354 pKa = 4.2EE355 pKa = 4.63TSQTEE360 pKa = 4.1GG361 pKa = 3.22
Molecular weight: 41.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4GZI9|Q4GZI9_9GEMI Protein V2 OS=Alternanthera yellow vein virus OX=337826 GN=av2 PE=3 SV=1
MM1 pKa = 7.6SKK3 pKa = 10.24RR4 pKa = 11.84AADD7 pKa = 3.97MIISSSGSRR16 pKa = 11.84VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFDD24 pKa = 3.06SPMARR29 pKa = 11.84RR30 pKa = 11.84ATAPIVRR37 pKa = 11.84ATRR40 pKa = 11.84KK41 pKa = 7.64QQWSSRR47 pKa = 11.84PMYY50 pKa = 10.05RR51 pKa = 11.84KK52 pKa = 8.59PRR54 pKa = 11.84LYY56 pKa = 10.8RR57 pKa = 11.84MYY59 pKa = 10.37RR60 pKa = 11.84SPDD63 pKa = 3.16VPYY66 pKa = 10.77GCEE69 pKa = 4.42GPCKK73 pKa = 10.15VQSFEE78 pKa = 4.21KK79 pKa = 10.35KK80 pKa = 9.61HH81 pKa = 6.23DD82 pKa = 3.73VSHH85 pKa = 6.9TGTLLCVSDD94 pKa = 3.64VTRR97 pKa = 11.84GSGITHH103 pKa = 6.35RR104 pKa = 11.84VGKK107 pKa = 9.68RR108 pKa = 11.84FCIKK112 pKa = 10.14SIYY115 pKa = 9.87IIGKK119 pKa = 8.24IWMDD123 pKa = 3.52EE124 pKa = 4.09NIKK127 pKa = 10.71SKK129 pKa = 10.61NHH131 pKa = 5.85TNTVMFWLVRR141 pKa = 11.84DD142 pKa = 3.99RR143 pKa = 11.84RR144 pKa = 11.84PVTTPYY150 pKa = 11.3GFGEE154 pKa = 4.51AFNMYY159 pKa = 10.55DD160 pKa = 4.21NEE162 pKa = 4.43PSTATIKK169 pKa = 10.93NDD171 pKa = 3.08LRR173 pKa = 11.84DD174 pKa = 3.68RR175 pKa = 11.84LQVLHH180 pKa = 6.91RR181 pKa = 11.84FSATVTGGQYY191 pKa = 10.85ASKK194 pKa = 8.76EE195 pKa = 3.54QAMIKK200 pKa = 10.19RR201 pKa = 11.84FWRR204 pKa = 11.84INNHH208 pKa = 4.03VVYY211 pKa = 10.65NHH213 pKa = 5.67QEE215 pKa = 3.37KK216 pKa = 10.24AAYY219 pKa = 8.25EE220 pKa = 4.07NHH222 pKa = 6.29TEE224 pKa = 4.02NALVLYY230 pKa = 7.23MACTHH235 pKa = 7.04ASNPVYY241 pKa = 9.86ATLKK245 pKa = 9.47IRR247 pKa = 11.84IYY249 pKa = 10.66FYY251 pKa = 11.18DD252 pKa = 3.06SVMNN256 pKa = 4.52
MM1 pKa = 7.6SKK3 pKa = 10.24RR4 pKa = 11.84AADD7 pKa = 3.97MIISSSGSRR16 pKa = 11.84VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LNFDD24 pKa = 3.06SPMARR29 pKa = 11.84RR30 pKa = 11.84ATAPIVRR37 pKa = 11.84ATRR40 pKa = 11.84KK41 pKa = 7.64QQWSSRR47 pKa = 11.84PMYY50 pKa = 10.05RR51 pKa = 11.84KK52 pKa = 8.59PRR54 pKa = 11.84LYY56 pKa = 10.8RR57 pKa = 11.84MYY59 pKa = 10.37RR60 pKa = 11.84SPDD63 pKa = 3.16VPYY66 pKa = 10.77GCEE69 pKa = 4.42GPCKK73 pKa = 10.15VQSFEE78 pKa = 4.21KK79 pKa = 10.35KK80 pKa = 9.61HH81 pKa = 6.23DD82 pKa = 3.73VSHH85 pKa = 6.9TGTLLCVSDD94 pKa = 3.64VTRR97 pKa = 11.84GSGITHH103 pKa = 6.35RR104 pKa = 11.84VGKK107 pKa = 9.68RR108 pKa = 11.84FCIKK112 pKa = 10.14SIYY115 pKa = 9.87IIGKK119 pKa = 8.24IWMDD123 pKa = 3.52EE124 pKa = 4.09NIKK127 pKa = 10.71SKK129 pKa = 10.61NHH131 pKa = 5.85TNTVMFWLVRR141 pKa = 11.84DD142 pKa = 3.99RR143 pKa = 11.84RR144 pKa = 11.84PVTTPYY150 pKa = 11.3GFGEE154 pKa = 4.51AFNMYY159 pKa = 10.55DD160 pKa = 4.21NEE162 pKa = 4.43PSTATIKK169 pKa = 10.93NDD171 pKa = 3.08LRR173 pKa = 11.84DD174 pKa = 3.68RR175 pKa = 11.84LQVLHH180 pKa = 6.91RR181 pKa = 11.84FSATVTGGQYY191 pKa = 10.85ASKK194 pKa = 8.76EE195 pKa = 3.54QAMIKK200 pKa = 10.19RR201 pKa = 11.84FWRR204 pKa = 11.84INNHH208 pKa = 4.03VVYY211 pKa = 10.65NHH213 pKa = 5.67QEE215 pKa = 3.37KK216 pKa = 10.24AAYY219 pKa = 8.25EE220 pKa = 4.07NHH222 pKa = 6.29TEE224 pKa = 4.02NALVLYY230 pKa = 7.23MACTHH235 pKa = 7.04ASNPVYY241 pKa = 9.86ATLKK245 pKa = 9.47IRR247 pKa = 11.84IYY249 pKa = 10.66FYY251 pKa = 11.18DD252 pKa = 3.06SVMNN256 pKa = 4.52
Molecular weight: 29.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1096 |
96 |
361 |
182.7 |
20.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.657 ± 0.79 | 2.372 ± 0.484 |
4.927 ± 0.613 | 4.471 ± 0.825 |
4.015 ± 0.664 | 4.745 ± 0.597 |
3.65 ± 0.513 | 6.387 ± 0.836 |
4.927 ± 0.667 | 7.117 ± 1.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.555 ± 0.71 | 5.566 ± 0.872 |
5.748 ± 0.827 | 5.292 ± 0.688 |
7.208 ± 1.118 | 7.573 ± 1.136 |
6.843 ± 1.191 | 5.292 ± 0.988 |
1.46 ± 0.268 | 4.197 ± 0.487 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
