
bacterium HR35
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
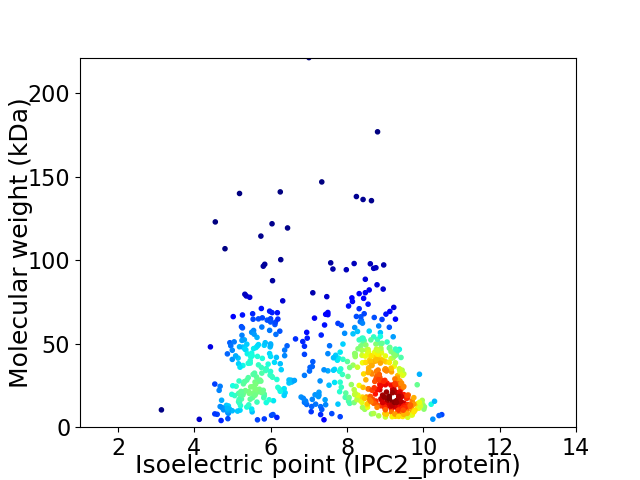
Virtual 2D-PAGE plot for 609 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H6A8W0|A0A2H6A8W0_9BACT Methionine aminopeptidase OS=bacterium HR35 OX=2035430 GN=map PE=3 SV=1
MM1 pKa = 7.5KK2 pKa = 10.19KK3 pKa = 9.58NHH5 pKa = 7.0PKK7 pKa = 10.61LSTRR11 pKa = 11.84FYY13 pKa = 10.88LLSTRR18 pKa = 11.84FLVLLLLISMFFFGYY33 pKa = 9.17PVIWKK38 pKa = 9.54NPRR41 pKa = 11.84IPPKK45 pKa = 10.29IKK47 pKa = 9.69EE48 pKa = 4.01AQAAISFIGSVEE60 pKa = 3.82GSAANGGDD68 pKa = 3.5VTLSLSGLGLQEE80 pKa = 4.2GDD82 pKa = 3.95LVIVAYY88 pKa = 10.34AIGDD92 pKa = 3.57NDD94 pKa = 3.22NVNFNMAILSPAGYY108 pKa = 8.62TEE110 pKa = 4.19VADD113 pKa = 3.97LHH115 pKa = 8.01SDD117 pKa = 3.44DD118 pKa = 4.6TQDD121 pKa = 3.33VDD123 pKa = 5.24LGVYY127 pKa = 8.15WKK129 pKa = 11.11VMGATPDD136 pKa = 3.29TSVTVDD142 pKa = 3.33GLGGTDD148 pKa = 3.14ASVAAVAMAFRR159 pKa = 11.84GVDD162 pKa = 3.21PATPMDD168 pKa = 4.89VIPTTATGINTMHH181 pKa = 7.44PNPPAIDD188 pKa = 3.78YY189 pKa = 10.59VSTGVWTVIAGASGHH204 pKa = 6.54ALNLANTNNPYY215 pKa = 9.38TFPSGYY221 pKa = 7.31TTDD224 pKa = 3.25NVQRR228 pKa = 11.84PEE230 pKa = 4.06NDD232 pKa = 3.24TSDD235 pKa = 3.53VTVGLGYY242 pKa = 8.84RR243 pKa = 11.84TNPADD248 pKa = 3.91PEE250 pKa = 4.54DD251 pKa = 4.53PGVMTHH257 pKa = 6.58SGTDD261 pKa = 3.29STSYY265 pKa = 10.22SWAAVTIALRR275 pKa = 11.84PKK277 pKa = 10.13PPSTFNQSAYY287 pKa = 10.85RR288 pKa = 11.84FFNNLDD294 pKa = 3.62STDD297 pKa = 3.55VGSPLANQNTPASLTSAGQAFRR319 pKa = 11.84LRR321 pKa = 11.84LLIHH325 pKa = 6.52VGTSNLLEE333 pKa = 4.32NEE335 pKa = 4.15QSFKK339 pKa = 11.28LQFGTSTGSSCTSSPPASWADD360 pKa = 3.49VTTSTPIAFNDD371 pKa = 3.55NPTPADD377 pKa = 3.72GATLTANTNDD387 pKa = 3.77PTHH390 pKa = 6.37GADD393 pKa = 3.23IIVNQTYY400 pKa = 10.56EE401 pKa = 4.04EE402 pKa = 4.65LNNFTNSVSSINAGEE417 pKa = 4.09DD418 pKa = 3.76GKK420 pKa = 10.3WDD422 pKa = 4.48FSLKK426 pKa = 10.98DD427 pKa = 3.28NGAPAGTTYY436 pKa = 10.91CLRR439 pKa = 11.84VVKK442 pKa = 10.72ADD444 pKa = 3.34GTLLDD449 pKa = 4.22TYY451 pKa = 10.98SVIPQITTAAAPILSAYY468 pKa = 10.44RR469 pKa = 11.84FFNNLDD475 pKa = 3.62STDD478 pKa = 3.55VGSPLANQDD487 pKa = 2.98TAATLTSAGQAFRR500 pKa = 11.84LRR502 pKa = 11.84LLLHH506 pKa = 6.64LPSGLNFNSPTSSNNLIEE524 pKa = 4.89SGLIPAGSGGGRR536 pKa = 11.84IRR538 pKa = 11.84IAAPTPNRR546 pKa = 11.84LYY548 pKa = 10.81LVYY551 pKa = 11.0YY552 pKa = 11.02DD553 pKa = 4.02MTNLDD558 pKa = 3.86LRR560 pKa = 11.84FCRR563 pKa = 11.84STNGGVSWSCNAIEE577 pKa = 4.32STGDD581 pKa = 3.24VGVTPDD587 pKa = 4.66IYY589 pKa = 11.42ALDD592 pKa = 3.97EE593 pKa = 4.26NNIFITHH600 pKa = 6.94RR601 pKa = 11.84DD602 pKa = 3.32ATNGNLRR609 pKa = 11.84FCKK612 pKa = 10.29SSNGGLNFSCSILEE626 pKa = 4.39AGWQGFDD633 pKa = 3.68SSIKK637 pKa = 10.44AISTSTIFVSYY648 pKa = 10.81APAPANFSYY657 pKa = 10.77SILRR661 pKa = 11.84FCRR664 pKa = 11.84TTDD667 pKa = 4.37GGNTWNCSDD676 pKa = 3.96VEE678 pKa = 4.61GNTTTQYY685 pKa = 10.9GWYY688 pKa = 9.52SSLDD692 pKa = 3.41AVDD695 pKa = 4.11SNNIYY700 pKa = 9.49IAHH703 pKa = 6.64YY704 pKa = 10.68DD705 pKa = 4.06GINLDD710 pKa = 3.88LRR712 pKa = 11.84FCRR715 pKa = 11.84TSNGGTTWNCGVIEE729 pKa = 4.26TTGDD733 pKa = 2.97IGRR736 pKa = 11.84KK737 pKa = 8.77PSIKK741 pKa = 10.43AFNNNYY747 pKa = 9.82IYY749 pKa = 10.24IAHH752 pKa = 7.31RR753 pKa = 11.84DD754 pKa = 3.47EE755 pKa = 4.75TNNQLRR761 pKa = 11.84FCRR764 pKa = 11.84TTDD767 pKa = 4.24GGTSWNCNAIEE778 pKa = 3.98IGGFSSSLKK787 pKa = 10.42IVDD790 pKa = 3.53KK791 pKa = 10.78DD792 pKa = 3.66VVYY795 pKa = 10.39IAHH798 pKa = 7.18FDD800 pKa = 3.53FTNASIRR807 pKa = 11.84LCNTNNGGQTWSCSTIANAPPTLTSGEE834 pKa = 4.19NLSLSLDD841 pKa = 3.66LLDD844 pKa = 4.96PYY846 pKa = 9.99MIYY849 pKa = 10.38VAYY852 pKa = 10.29GFTTDD857 pKa = 2.84SSNEE861 pKa = 3.56VRR863 pKa = 11.84LYY865 pKa = 10.75KK866 pKa = 10.04YY867 pKa = 9.07TGNDD871 pKa = 3.25FFKK874 pKa = 10.76LQYY877 pKa = 10.83SVMSGTCDD885 pKa = 2.86TSFSGEE891 pKa = 4.09VYY893 pKa = 10.82NDD895 pKa = 3.11VGFDD899 pKa = 3.58TPIAFYY905 pKa = 11.43NNNTTSDD912 pKa = 3.35ASTPTTNTNDD922 pKa = 3.65PTHH925 pKa = 5.84GTDD928 pKa = 3.28TIVYY932 pKa = 7.19QTYY935 pKa = 9.81KK936 pKa = 10.09EE937 pKa = 4.28KK938 pKa = 11.04KK939 pKa = 9.12FFTNRR944 pKa = 11.84TNIFSGSDD952 pKa = 3.61GLWDD956 pKa = 4.63FVLRR960 pKa = 11.84DD961 pKa = 3.68NNAPSNTTYY970 pKa = 11.0CLRR973 pKa = 11.84VVKK976 pKa = 10.7ADD978 pKa = 3.32GSLLQGYY985 pKa = 7.97NVIPQITTAAAAPITCTFSATSTSFSSLSPSAVSTSSPDD1024 pKa = 2.9ITITVTSSAGFIISVNDD1041 pKa = 3.73AGNGTNPGLYY1051 pKa = 9.97KK1052 pKa = 9.93STSPTYY1058 pKa = 10.58LIPSPNSSYY1067 pKa = 10.65TATATLVAGVDD1078 pKa = 4.06GYY1080 pKa = 11.39GIQATTTNSNISINPRR1096 pKa = 11.84YY1097 pKa = 9.19NVSGDD1102 pKa = 3.6TVGGLTTTTITLASSSVAVSSAQITIKK1129 pKa = 10.61HH1130 pKa = 5.51KK1131 pKa = 10.75AAVSSLAPTGNYY1143 pKa = 9.55QDD1145 pKa = 4.51TITYY1149 pKa = 10.21SCTSPP1154 pKa = 3.49
MM1 pKa = 7.5KK2 pKa = 10.19KK3 pKa = 9.58NHH5 pKa = 7.0PKK7 pKa = 10.61LSTRR11 pKa = 11.84FYY13 pKa = 10.88LLSTRR18 pKa = 11.84FLVLLLLISMFFFGYY33 pKa = 9.17PVIWKK38 pKa = 9.54NPRR41 pKa = 11.84IPPKK45 pKa = 10.29IKK47 pKa = 9.69EE48 pKa = 4.01AQAAISFIGSVEE60 pKa = 3.82GSAANGGDD68 pKa = 3.5VTLSLSGLGLQEE80 pKa = 4.2GDD82 pKa = 3.95LVIVAYY88 pKa = 10.34AIGDD92 pKa = 3.57NDD94 pKa = 3.22NVNFNMAILSPAGYY108 pKa = 8.62TEE110 pKa = 4.19VADD113 pKa = 3.97LHH115 pKa = 8.01SDD117 pKa = 3.44DD118 pKa = 4.6TQDD121 pKa = 3.33VDD123 pKa = 5.24LGVYY127 pKa = 8.15WKK129 pKa = 11.11VMGATPDD136 pKa = 3.29TSVTVDD142 pKa = 3.33GLGGTDD148 pKa = 3.14ASVAAVAMAFRR159 pKa = 11.84GVDD162 pKa = 3.21PATPMDD168 pKa = 4.89VIPTTATGINTMHH181 pKa = 7.44PNPPAIDD188 pKa = 3.78YY189 pKa = 10.59VSTGVWTVIAGASGHH204 pKa = 6.54ALNLANTNNPYY215 pKa = 9.38TFPSGYY221 pKa = 7.31TTDD224 pKa = 3.25NVQRR228 pKa = 11.84PEE230 pKa = 4.06NDD232 pKa = 3.24TSDD235 pKa = 3.53VTVGLGYY242 pKa = 8.84RR243 pKa = 11.84TNPADD248 pKa = 3.91PEE250 pKa = 4.54DD251 pKa = 4.53PGVMTHH257 pKa = 6.58SGTDD261 pKa = 3.29STSYY265 pKa = 10.22SWAAVTIALRR275 pKa = 11.84PKK277 pKa = 10.13PPSTFNQSAYY287 pKa = 10.85RR288 pKa = 11.84FFNNLDD294 pKa = 3.62STDD297 pKa = 3.55VGSPLANQNTPASLTSAGQAFRR319 pKa = 11.84LRR321 pKa = 11.84LLIHH325 pKa = 6.52VGTSNLLEE333 pKa = 4.32NEE335 pKa = 4.15QSFKK339 pKa = 11.28LQFGTSTGSSCTSSPPASWADD360 pKa = 3.49VTTSTPIAFNDD371 pKa = 3.55NPTPADD377 pKa = 3.72GATLTANTNDD387 pKa = 3.77PTHH390 pKa = 6.37GADD393 pKa = 3.23IIVNQTYY400 pKa = 10.56EE401 pKa = 4.04EE402 pKa = 4.65LNNFTNSVSSINAGEE417 pKa = 4.09DD418 pKa = 3.76GKK420 pKa = 10.3WDD422 pKa = 4.48FSLKK426 pKa = 10.98DD427 pKa = 3.28NGAPAGTTYY436 pKa = 10.91CLRR439 pKa = 11.84VVKK442 pKa = 10.72ADD444 pKa = 3.34GTLLDD449 pKa = 4.22TYY451 pKa = 10.98SVIPQITTAAAPILSAYY468 pKa = 10.44RR469 pKa = 11.84FFNNLDD475 pKa = 3.62STDD478 pKa = 3.55VGSPLANQDD487 pKa = 2.98TAATLTSAGQAFRR500 pKa = 11.84LRR502 pKa = 11.84LLLHH506 pKa = 6.64LPSGLNFNSPTSSNNLIEE524 pKa = 4.89SGLIPAGSGGGRR536 pKa = 11.84IRR538 pKa = 11.84IAAPTPNRR546 pKa = 11.84LYY548 pKa = 10.81LVYY551 pKa = 11.0YY552 pKa = 11.02DD553 pKa = 4.02MTNLDD558 pKa = 3.86LRR560 pKa = 11.84FCRR563 pKa = 11.84STNGGVSWSCNAIEE577 pKa = 4.32STGDD581 pKa = 3.24VGVTPDD587 pKa = 4.66IYY589 pKa = 11.42ALDD592 pKa = 3.97EE593 pKa = 4.26NNIFITHH600 pKa = 6.94RR601 pKa = 11.84DD602 pKa = 3.32ATNGNLRR609 pKa = 11.84FCKK612 pKa = 10.29SSNGGLNFSCSILEE626 pKa = 4.39AGWQGFDD633 pKa = 3.68SSIKK637 pKa = 10.44AISTSTIFVSYY648 pKa = 10.81APAPANFSYY657 pKa = 10.77SILRR661 pKa = 11.84FCRR664 pKa = 11.84TTDD667 pKa = 4.37GGNTWNCSDD676 pKa = 3.96VEE678 pKa = 4.61GNTTTQYY685 pKa = 10.9GWYY688 pKa = 9.52SSLDD692 pKa = 3.41AVDD695 pKa = 4.11SNNIYY700 pKa = 9.49IAHH703 pKa = 6.64YY704 pKa = 10.68DD705 pKa = 4.06GINLDD710 pKa = 3.88LRR712 pKa = 11.84FCRR715 pKa = 11.84TSNGGTTWNCGVIEE729 pKa = 4.26TTGDD733 pKa = 2.97IGRR736 pKa = 11.84KK737 pKa = 8.77PSIKK741 pKa = 10.43AFNNNYY747 pKa = 9.82IYY749 pKa = 10.24IAHH752 pKa = 7.31RR753 pKa = 11.84DD754 pKa = 3.47EE755 pKa = 4.75TNNQLRR761 pKa = 11.84FCRR764 pKa = 11.84TTDD767 pKa = 4.24GGTSWNCNAIEE778 pKa = 3.98IGGFSSSLKK787 pKa = 10.42IVDD790 pKa = 3.53KK791 pKa = 10.78DD792 pKa = 3.66VVYY795 pKa = 10.39IAHH798 pKa = 7.18FDD800 pKa = 3.53FTNASIRR807 pKa = 11.84LCNTNNGGQTWSCSTIANAPPTLTSGEE834 pKa = 4.19NLSLSLDD841 pKa = 3.66LLDD844 pKa = 4.96PYY846 pKa = 9.99MIYY849 pKa = 10.38VAYY852 pKa = 10.29GFTTDD857 pKa = 2.84SSNEE861 pKa = 3.56VRR863 pKa = 11.84LYY865 pKa = 10.75KK866 pKa = 10.04YY867 pKa = 9.07TGNDD871 pKa = 3.25FFKK874 pKa = 10.76LQYY877 pKa = 10.83SVMSGTCDD885 pKa = 2.86TSFSGEE891 pKa = 4.09VYY893 pKa = 10.82NDD895 pKa = 3.11VGFDD899 pKa = 3.58TPIAFYY905 pKa = 11.43NNNTTSDD912 pKa = 3.35ASTPTTNTNDD922 pKa = 3.65PTHH925 pKa = 5.84GTDD928 pKa = 3.28TIVYY932 pKa = 7.19QTYY935 pKa = 9.81KK936 pKa = 10.09EE937 pKa = 4.28KK938 pKa = 11.04KK939 pKa = 9.12FFTNRR944 pKa = 11.84TNIFSGSDD952 pKa = 3.61GLWDD956 pKa = 4.63FVLRR960 pKa = 11.84DD961 pKa = 3.68NNAPSNTTYY970 pKa = 11.0CLRR973 pKa = 11.84VVKK976 pKa = 10.7ADD978 pKa = 3.32GSLLQGYY985 pKa = 7.97NVIPQITTAAAAPITCTFSATSTSFSSLSPSAVSTSSPDD1024 pKa = 2.9ITITVTSSAGFIISVNDD1041 pKa = 3.73AGNGTNPGLYY1051 pKa = 9.97KK1052 pKa = 9.93STSPTYY1058 pKa = 10.58LIPSPNSSYY1067 pKa = 10.65TATATLVAGVDD1078 pKa = 4.06GYY1080 pKa = 11.39GIQATTTNSNISINPRR1096 pKa = 11.84YY1097 pKa = 9.19NVSGDD1102 pKa = 3.6TVGGLTTTTITLASSSVAVSSAQITIKK1129 pKa = 10.61HH1130 pKa = 5.51KK1131 pKa = 10.75AAVSSLAPTGNYY1143 pKa = 9.55QDD1145 pKa = 4.51TITYY1149 pKa = 10.21SCTSPP1154 pKa = 3.49
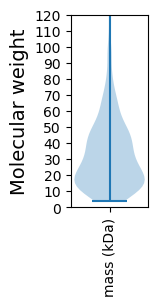
Molecular weight: 122.97 kDa
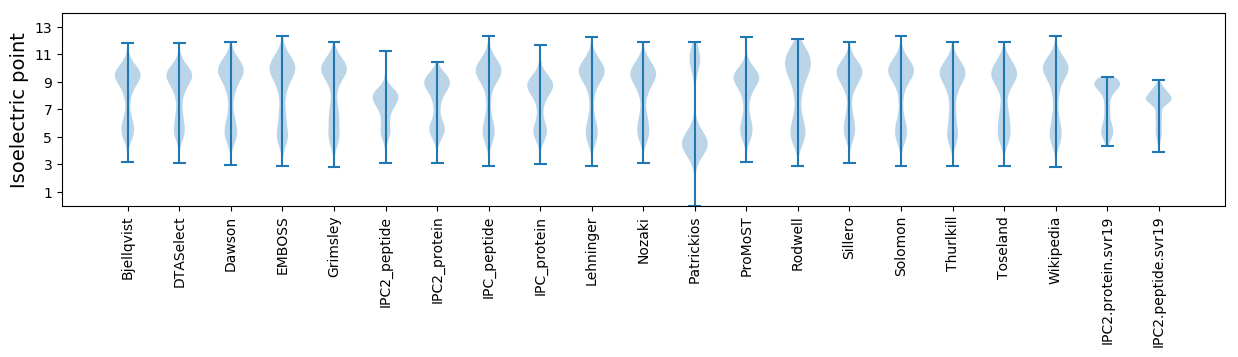
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H6A9N1|A0A2H6A9N1_9BACT Ribosome-recycling factor OS=bacterium HR35 OX=2035430 GN=frr PE=3 SV=1
MM1 pKa = 7.68PSKK4 pKa = 10.83AQIEE8 pKa = 4.37RR9 pKa = 11.84WKK11 pKa = 10.74KK12 pKa = 8.32EE13 pKa = 3.94PKK15 pKa = 9.95FKK17 pKa = 9.65TRR19 pKa = 11.84KK20 pKa = 9.33RR21 pKa = 11.84NFCFRR26 pKa = 11.84CGRR29 pKa = 11.84TRR31 pKa = 11.84GYY33 pKa = 10.73FRR35 pKa = 11.84DD36 pKa = 3.85FNLCRR41 pKa = 11.84ICLRR45 pKa = 11.84EE46 pKa = 3.65MAHH49 pKa = 6.67RR50 pKa = 11.84GEE52 pKa = 4.19IPGLKK57 pKa = 9.06KK58 pKa = 10.87ASWW61 pKa = 3.28
MM1 pKa = 7.68PSKK4 pKa = 10.83AQIEE8 pKa = 4.37RR9 pKa = 11.84WKK11 pKa = 10.74KK12 pKa = 8.32EE13 pKa = 3.94PKK15 pKa = 9.95FKK17 pKa = 9.65TRR19 pKa = 11.84KK20 pKa = 9.33RR21 pKa = 11.84NFCFRR26 pKa = 11.84CGRR29 pKa = 11.84TRR31 pKa = 11.84GYY33 pKa = 10.73FRR35 pKa = 11.84DD36 pKa = 3.85FNLCRR41 pKa = 11.84ICLRR45 pKa = 11.84EE46 pKa = 3.65MAHH49 pKa = 6.67RR50 pKa = 11.84GEE52 pKa = 4.19IPGLKK57 pKa = 9.06KK58 pKa = 10.87ASWW61 pKa = 3.28
Molecular weight: 7.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
183829 |
36 |
2003 |
301.9 |
34.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.555 ± 0.085 | 0.687 ± 0.031 |
4.198 ± 0.062 | 8.174 ± 0.147 |
6.507 ± 0.127 | 5.476 ± 0.089 |
1.19 ± 0.04 | 9.57 ± 0.1 |
10.57 ± 0.18 | 11.53 ± 0.145 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.233 ± 0.03 | 5.792 ± 0.097 |
3.803 ± 0.065 | 2.853 ± 0.052 |
3.825 ± 0.07 | 5.557 ± 0.128 |
4.16 ± 0.113 | 5.153 ± 0.077 |
0.943 ± 0.036 | 4.223 ± 0.078 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
