
Zamilon virus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; Sputnikvirus; Mimivirus-dependent virus Zamilon
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
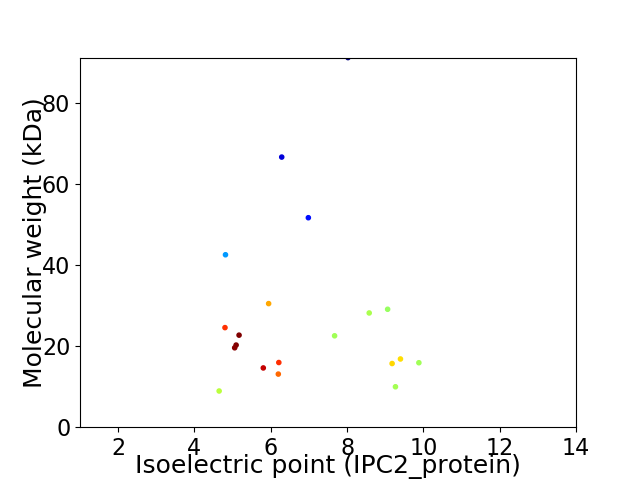
Virtual 2D-PAGE plot for 20 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|V6BPN4|V6BPN4_9VIRU Uncharacterized protein OS=Zamilon virus OX=1411887 GN=za3_9 PE=4 SV=1
MM1 pKa = 7.42SHH3 pKa = 6.54NCQNSNEE10 pKa = 4.38PDD12 pKa = 3.23TVYY15 pKa = 11.19YY16 pKa = 10.49DD17 pKa = 3.2IVIPYY22 pKa = 9.26NPNDD26 pKa = 3.92SGLSPATFQAQLTQPLLYY44 pKa = 10.98NPDD47 pKa = 3.41KK48 pKa = 11.12YY49 pKa = 10.92YY50 pKa = 11.11LSVVRR55 pKa = 11.84FSIPTQYY62 pKa = 10.36VPLMIPEE69 pKa = 4.44IQPFPNTNVNNTIYY83 pKa = 10.79SVTLEE88 pKa = 4.1YY89 pKa = 11.23NGVFSSQTFVQYY101 pKa = 10.7DD102 pKa = 3.56VSLTNPNDD110 pKa = 3.96TPPPPPTINNRR121 pKa = 11.84TVEE124 pKa = 4.2PTAYY128 pKa = 9.41YY129 pKa = 10.06YY130 pKa = 11.07VYY132 pKa = 10.58NFSPFLQMINKK143 pKa = 9.18ALSDD147 pKa = 3.94AFTEE151 pKa = 3.77ITMPVGAVAPYY162 pKa = 9.61FVYY165 pKa = 10.68SPVTQRR171 pKa = 11.84ISLVAQRR178 pKa = 11.84QFYY181 pKa = 10.47DD182 pKa = 3.46RR183 pKa = 11.84NLAQPIRR190 pKa = 11.84IYY192 pKa = 10.89CNEE195 pKa = 3.82ALFPFLDD202 pKa = 5.36GIPFGGLDD210 pKa = 3.69FNSVDD215 pKa = 4.1GRR217 pKa = 11.84DD218 pKa = 3.03ILFNVEE224 pKa = 3.9NLGNNLVQNQLTAPAYY240 pKa = 8.81PPEE243 pKa = 5.07FIQMEE248 pKa = 4.28QEE250 pKa = 4.21YY251 pKa = 8.58ATLSNWNAIKK261 pKa = 9.78TIQLVSNLLPINRR274 pKa = 11.84EE275 pKa = 4.06FIPSFRR281 pKa = 11.84NTNVGVVNSQGILADD296 pKa = 4.07FVPLVTLGPEE306 pKa = 3.77SRR308 pKa = 11.84TSIDD312 pKa = 3.43YY313 pKa = 9.88VANGPWRR320 pKa = 11.84LIDD323 pKa = 3.48MFGGVPITMVDD334 pKa = 3.81LSVYY338 pKa = 8.26WTDD341 pKa = 3.25QIGRR345 pKa = 11.84RR346 pKa = 11.84FVLDD350 pKa = 3.37VPRR353 pKa = 11.84GRR355 pKa = 11.84IATCKK360 pKa = 10.53LIFIKK365 pKa = 10.4KK366 pKa = 9.69DD367 pKa = 3.48LAGHH371 pKa = 5.1TLSRR375 pKa = 11.84KK376 pKa = 9.32
MM1 pKa = 7.42SHH3 pKa = 6.54NCQNSNEE10 pKa = 4.38PDD12 pKa = 3.23TVYY15 pKa = 11.19YY16 pKa = 10.49DD17 pKa = 3.2IVIPYY22 pKa = 9.26NPNDD26 pKa = 3.92SGLSPATFQAQLTQPLLYY44 pKa = 10.98NPDD47 pKa = 3.41KK48 pKa = 11.12YY49 pKa = 10.92YY50 pKa = 11.11LSVVRR55 pKa = 11.84FSIPTQYY62 pKa = 10.36VPLMIPEE69 pKa = 4.44IQPFPNTNVNNTIYY83 pKa = 10.79SVTLEE88 pKa = 4.1YY89 pKa = 11.23NGVFSSQTFVQYY101 pKa = 10.7DD102 pKa = 3.56VSLTNPNDD110 pKa = 3.96TPPPPPTINNRR121 pKa = 11.84TVEE124 pKa = 4.2PTAYY128 pKa = 9.41YY129 pKa = 10.06YY130 pKa = 11.07VYY132 pKa = 10.58NFSPFLQMINKK143 pKa = 9.18ALSDD147 pKa = 3.94AFTEE151 pKa = 3.77ITMPVGAVAPYY162 pKa = 9.61FVYY165 pKa = 10.68SPVTQRR171 pKa = 11.84ISLVAQRR178 pKa = 11.84QFYY181 pKa = 10.47DD182 pKa = 3.46RR183 pKa = 11.84NLAQPIRR190 pKa = 11.84IYY192 pKa = 10.89CNEE195 pKa = 3.82ALFPFLDD202 pKa = 5.36GIPFGGLDD210 pKa = 3.69FNSVDD215 pKa = 4.1GRR217 pKa = 11.84DD218 pKa = 3.03ILFNVEE224 pKa = 3.9NLGNNLVQNQLTAPAYY240 pKa = 8.81PPEE243 pKa = 5.07FIQMEE248 pKa = 4.28QEE250 pKa = 4.21YY251 pKa = 8.58ATLSNWNAIKK261 pKa = 9.78TIQLVSNLLPINRR274 pKa = 11.84EE275 pKa = 4.06FIPSFRR281 pKa = 11.84NTNVGVVNSQGILADD296 pKa = 4.07FVPLVTLGPEE306 pKa = 3.77SRR308 pKa = 11.84TSIDD312 pKa = 3.43YY313 pKa = 9.88VANGPWRR320 pKa = 11.84LIDD323 pKa = 3.48MFGGVPITMVDD334 pKa = 3.81LSVYY338 pKa = 8.26WTDD341 pKa = 3.25QIGRR345 pKa = 11.84RR346 pKa = 11.84FVLDD350 pKa = 3.37VPRR353 pKa = 11.84GRR355 pKa = 11.84IATCKK360 pKa = 10.53LIFIKK365 pKa = 10.4KK366 pKa = 9.69DD367 pKa = 3.48LAGHH371 pKa = 5.1TLSRR375 pKa = 11.84KK376 pKa = 9.32
Molecular weight: 42.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V6BPP1|V6BPP1_9VIRU DNA packaging ATPase OS=Zamilon virus OX=1411887 GN=za3_4 PE=4 SV=1
MM1 pKa = 7.35SKK3 pKa = 10.49KK4 pKa = 10.49VLLSGTAPRR13 pKa = 11.84GGCFCPNCGTEE24 pKa = 3.98VGGVLAGVLSGGRR37 pKa = 11.84TVKK40 pKa = 10.4RR41 pKa = 11.84RR42 pKa = 11.84VKK44 pKa = 10.5KK45 pKa = 10.83GGIGTKK51 pKa = 10.28AGAKK55 pKa = 9.16KK56 pKa = 10.45SPWVAFLQKK65 pKa = 9.93FAKK68 pKa = 8.6EE69 pKa = 3.84HH70 pKa = 4.73GMKK73 pKa = 10.37YY74 pKa = 10.19GEE76 pKa = 4.55AMQSPKK82 pKa = 10.82AKK84 pKa = 9.88KK85 pKa = 9.9QYY87 pKa = 9.72EE88 pKa = 4.06ALKK91 pKa = 10.03KK92 pKa = 10.65KK93 pKa = 10.3SGKK96 pKa = 4.66TTKK99 pKa = 10.16KK100 pKa = 9.94KK101 pKa = 10.77SSGSKK106 pKa = 7.97TKK108 pKa = 10.51KK109 pKa = 7.76ITKK112 pKa = 9.42KK113 pKa = 10.65KK114 pKa = 9.34PVKK117 pKa = 10.09KK118 pKa = 10.03SLKK121 pKa = 9.94GGSKK125 pKa = 8.62RR126 pKa = 11.84TKK128 pKa = 10.07RR129 pKa = 11.84SPKK132 pKa = 9.47MSNNDD137 pKa = 3.13QRR139 pKa = 11.84ILEE142 pKa = 4.3EE143 pKa = 4.17LMNMM147 pKa = 4.51
MM1 pKa = 7.35SKK3 pKa = 10.49KK4 pKa = 10.49VLLSGTAPRR13 pKa = 11.84GGCFCPNCGTEE24 pKa = 3.98VGGVLAGVLSGGRR37 pKa = 11.84TVKK40 pKa = 10.4RR41 pKa = 11.84RR42 pKa = 11.84VKK44 pKa = 10.5KK45 pKa = 10.83GGIGTKK51 pKa = 10.28AGAKK55 pKa = 9.16KK56 pKa = 10.45SPWVAFLQKK65 pKa = 9.93FAKK68 pKa = 8.6EE69 pKa = 3.84HH70 pKa = 4.73GMKK73 pKa = 10.37YY74 pKa = 10.19GEE76 pKa = 4.55AMQSPKK82 pKa = 10.82AKK84 pKa = 9.88KK85 pKa = 9.9QYY87 pKa = 9.72EE88 pKa = 4.06ALKK91 pKa = 10.03KK92 pKa = 10.65KK93 pKa = 10.3SGKK96 pKa = 4.66TTKK99 pKa = 10.16KK100 pKa = 9.94KK101 pKa = 10.77SSGSKK106 pKa = 7.97TKK108 pKa = 10.51KK109 pKa = 7.76ITKK112 pKa = 9.42KK113 pKa = 10.65KK114 pKa = 9.34PVKK117 pKa = 10.09KK118 pKa = 10.03SLKK121 pKa = 9.94GGSKK125 pKa = 8.62RR126 pKa = 11.84TKK128 pKa = 10.07RR129 pKa = 11.84SPKK132 pKa = 9.47MSNNDD137 pKa = 3.13QRR139 pKa = 11.84ILEE142 pKa = 4.3EE143 pKa = 4.17LMNMM147 pKa = 4.51
Molecular weight: 15.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4928 |
73 |
778 |
246.4 |
28.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.485 ± 0.379 | 1.157 ± 0.221 |
5.641 ± 0.482 | 6.088 ± 0.764 |
4.282 ± 0.403 | 6.189 ± 1.109 |
1.238 ± 0.151 | 7.325 ± 0.476 |
10.329 ± 1.358 | 8.34 ± 0.348 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.597 ± 0.197 | 7.711 ± 0.464 |
4.282 ± 0.498 | 3.389 ± 0.428 |
3.369 ± 0.358 | 6.757 ± 0.588 |
5.662 ± 0.535 | 5.641 ± 0.394 |
0.609 ± 0.089 | 4.911 ± 0.403 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
