
Brachybacterium sp. SGAir0954
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Dermabacteraceae; Brachybacterium; unclassified Brachybacterium
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
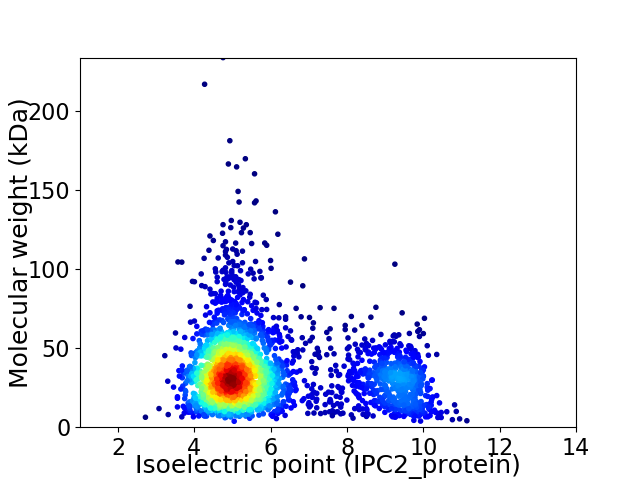
Virtual 2D-PAGE plot for 2953 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
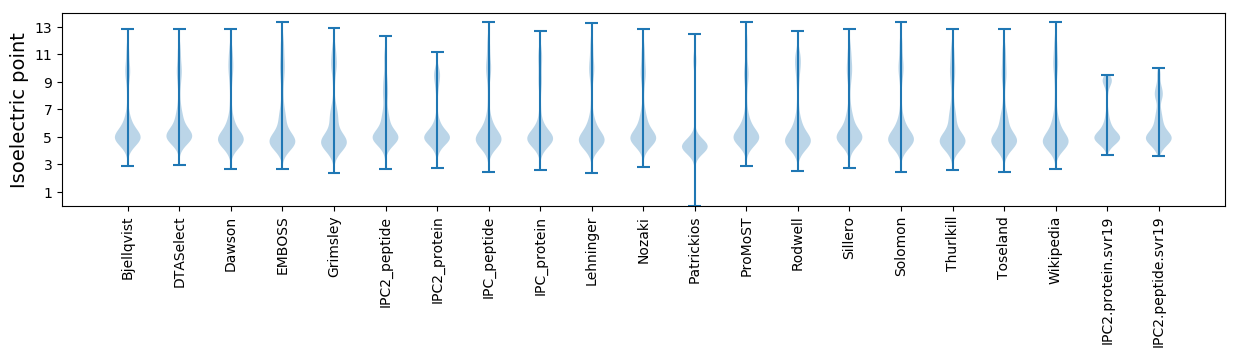
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8TH95|A0A4P8TH95_9MICO Oxidoreductase OS=Brachybacterium sp. SGAir0954 OX=2571029 GN=C1N80_06695 PE=4 SV=1
MM1 pKa = 7.65SSTLPASRR9 pKa = 11.84RR10 pKa = 11.84PLARR14 pKa = 11.84LGVLAAAALLAVTGCTPGGGSDD36 pKa = 4.23GADD39 pKa = 3.37ASDD42 pKa = 3.69GGADD46 pKa = 3.37QGAASTGTAAAEE58 pKa = 3.96LADD61 pKa = 4.91LGTSHH66 pKa = 7.43AADD69 pKa = 4.51LDD71 pKa = 3.79VDD73 pKa = 4.19PASDD77 pKa = 3.4PAYY80 pKa = 10.13AQYY83 pKa = 11.27YY84 pKa = 8.81DD85 pKa = 3.8QQIDD89 pKa = 2.99WGACEE94 pKa = 4.71DD95 pKa = 3.67VSGSVGLEE103 pKa = 3.96CGTLTVPLAWDD114 pKa = 4.25DD115 pKa = 4.01PSAGDD120 pKa = 3.67VEE122 pKa = 4.65LAVARR127 pKa = 11.84QGATVDD133 pKa = 3.42AKK135 pKa = 11.17GSLVVNPGGPGGSGVDD151 pKa = 3.48MAAQADD157 pKa = 4.35AYY159 pKa = 10.83FSADD163 pKa = 2.79VMASYY168 pKa = 11.06DD169 pKa = 3.58VVGFDD174 pKa = 3.51PRR176 pKa = 11.84GVARR180 pKa = 11.84SQGIDD185 pKa = 3.45CLTDD189 pKa = 3.24EE190 pKa = 6.09DD191 pKa = 5.03LDD193 pKa = 3.91QMRR196 pKa = 11.84ADD198 pKa = 4.05SFDD201 pKa = 4.12LLTEE205 pKa = 4.31GGEE208 pKa = 4.34RR209 pKa = 11.84AQHH212 pKa = 5.68PWVEE216 pKa = 4.39RR217 pKa = 11.84LAEE220 pKa = 4.32GCEE223 pKa = 4.05DD224 pKa = 3.92DD225 pKa = 4.76PGSAEE230 pKa = 4.33LLPYY234 pKa = 10.54LDD236 pKa = 4.1TASSARR242 pKa = 11.84DD243 pKa = 3.44LDD245 pKa = 3.85VLRR248 pKa = 11.84AAAGSEE254 pKa = 3.58QLDD257 pKa = 3.9YY258 pKa = 11.17FGYY261 pKa = 10.4SYY263 pKa = 10.69GTFLGSTYY271 pKa = 10.96AGLYY275 pKa = 7.78PEE277 pKa = 4.36RR278 pKa = 11.84VGRR281 pKa = 11.84MVLDD285 pKa = 3.99GAVDD289 pKa = 3.69PTLTADD295 pKa = 4.69DD296 pKa = 5.32FSAGQAGGFEE306 pKa = 4.16QATRR310 pKa = 11.84TFLEE314 pKa = 4.64DD315 pKa = 3.49CLKK318 pKa = 10.59QGEE321 pKa = 4.89GEE323 pKa = 4.47CPFTGDD329 pKa = 2.79VDD331 pKa = 4.21AAAQQLLDD339 pKa = 4.39LFEE342 pKa = 4.94KK343 pKa = 10.56AQSDD347 pKa = 4.31PLPTDD352 pKa = 3.87DD353 pKa = 5.3PDD355 pKa = 4.4RR356 pKa = 11.84PLTGALARR364 pKa = 11.84SAVLMLMYY372 pKa = 10.48DD373 pKa = 3.7DD374 pKa = 6.06SSWQYY379 pKa = 10.58GRR381 pKa = 11.84EE382 pKa = 4.24GIAAALDD389 pKa = 3.69GDD391 pKa = 4.3GQILLNIADD400 pKa = 3.66QSASRR405 pKa = 11.84NRR407 pKa = 11.84DD408 pKa = 2.43GSYY411 pKa = 10.51SGNGNEE417 pKa = 5.63AITAVNCLDD426 pKa = 3.76RR427 pKa = 11.84PGLASEE433 pKa = 4.48EE434 pKa = 4.41EE435 pKa = 4.45EE436 pKa = 3.99TALAHH441 pKa = 6.57QLAKK445 pKa = 10.49DD446 pKa = 3.73DD447 pKa = 4.53PVWGSSFSYY456 pKa = 10.72AATTCGYY463 pKa = 9.42WPEE466 pKa = 4.14KK467 pKa = 9.61PVRR470 pKa = 11.84EE471 pKa = 4.28PAPVTADD478 pKa = 3.1GAAPIVVIGTTGDD491 pKa = 3.39PATPYY496 pKa = 9.2TWAEE500 pKa = 3.98SLADD504 pKa = 3.72QLSSGVLLTFEE515 pKa = 5.22GNGHH519 pKa = 4.96TAYY522 pKa = 10.14GRR524 pKa = 11.84SGGCIEE530 pKa = 4.49EE531 pKa = 4.2QVDD534 pKa = 3.8AYY536 pKa = 10.85LLEE539 pKa = 4.55GTTPAEE545 pKa = 4.45GARR548 pKa = 11.84CC549 pKa = 3.48
MM1 pKa = 7.65SSTLPASRR9 pKa = 11.84RR10 pKa = 11.84PLARR14 pKa = 11.84LGVLAAAALLAVTGCTPGGGSDD36 pKa = 4.23GADD39 pKa = 3.37ASDD42 pKa = 3.69GGADD46 pKa = 3.37QGAASTGTAAAEE58 pKa = 3.96LADD61 pKa = 4.91LGTSHH66 pKa = 7.43AADD69 pKa = 4.51LDD71 pKa = 3.79VDD73 pKa = 4.19PASDD77 pKa = 3.4PAYY80 pKa = 10.13AQYY83 pKa = 11.27YY84 pKa = 8.81DD85 pKa = 3.8QQIDD89 pKa = 2.99WGACEE94 pKa = 4.71DD95 pKa = 3.67VSGSVGLEE103 pKa = 3.96CGTLTVPLAWDD114 pKa = 4.25DD115 pKa = 4.01PSAGDD120 pKa = 3.67VEE122 pKa = 4.65LAVARR127 pKa = 11.84QGATVDD133 pKa = 3.42AKK135 pKa = 11.17GSLVVNPGGPGGSGVDD151 pKa = 3.48MAAQADD157 pKa = 4.35AYY159 pKa = 10.83FSADD163 pKa = 2.79VMASYY168 pKa = 11.06DD169 pKa = 3.58VVGFDD174 pKa = 3.51PRR176 pKa = 11.84GVARR180 pKa = 11.84SQGIDD185 pKa = 3.45CLTDD189 pKa = 3.24EE190 pKa = 6.09DD191 pKa = 5.03LDD193 pKa = 3.91QMRR196 pKa = 11.84ADD198 pKa = 4.05SFDD201 pKa = 4.12LLTEE205 pKa = 4.31GGEE208 pKa = 4.34RR209 pKa = 11.84AQHH212 pKa = 5.68PWVEE216 pKa = 4.39RR217 pKa = 11.84LAEE220 pKa = 4.32GCEE223 pKa = 4.05DD224 pKa = 3.92DD225 pKa = 4.76PGSAEE230 pKa = 4.33LLPYY234 pKa = 10.54LDD236 pKa = 4.1TASSARR242 pKa = 11.84DD243 pKa = 3.44LDD245 pKa = 3.85VLRR248 pKa = 11.84AAAGSEE254 pKa = 3.58QLDD257 pKa = 3.9YY258 pKa = 11.17FGYY261 pKa = 10.4SYY263 pKa = 10.69GTFLGSTYY271 pKa = 10.96AGLYY275 pKa = 7.78PEE277 pKa = 4.36RR278 pKa = 11.84VGRR281 pKa = 11.84MVLDD285 pKa = 3.99GAVDD289 pKa = 3.69PTLTADD295 pKa = 4.69DD296 pKa = 5.32FSAGQAGGFEE306 pKa = 4.16QATRR310 pKa = 11.84TFLEE314 pKa = 4.64DD315 pKa = 3.49CLKK318 pKa = 10.59QGEE321 pKa = 4.89GEE323 pKa = 4.47CPFTGDD329 pKa = 2.79VDD331 pKa = 4.21AAAQQLLDD339 pKa = 4.39LFEE342 pKa = 4.94KK343 pKa = 10.56AQSDD347 pKa = 4.31PLPTDD352 pKa = 3.87DD353 pKa = 5.3PDD355 pKa = 4.4RR356 pKa = 11.84PLTGALARR364 pKa = 11.84SAVLMLMYY372 pKa = 10.48DD373 pKa = 3.7DD374 pKa = 6.06SSWQYY379 pKa = 10.58GRR381 pKa = 11.84EE382 pKa = 4.24GIAAALDD389 pKa = 3.69GDD391 pKa = 4.3GQILLNIADD400 pKa = 3.66QSASRR405 pKa = 11.84NRR407 pKa = 11.84DD408 pKa = 2.43GSYY411 pKa = 10.51SGNGNEE417 pKa = 5.63AITAVNCLDD426 pKa = 3.76RR427 pKa = 11.84PGLASEE433 pKa = 4.48EE434 pKa = 4.41EE435 pKa = 4.45EE436 pKa = 3.99TALAHH441 pKa = 6.57QLAKK445 pKa = 10.49DD446 pKa = 3.73DD447 pKa = 4.53PVWGSSFSYY456 pKa = 10.72AATTCGYY463 pKa = 9.42WPEE466 pKa = 4.14KK467 pKa = 9.61PVRR470 pKa = 11.84EE471 pKa = 4.28PAPVTADD478 pKa = 3.1GAAPIVVIGTTGDD491 pKa = 3.39PATPYY496 pKa = 9.2TWAEE500 pKa = 3.98SLADD504 pKa = 3.72QLSSGVLLTFEE515 pKa = 5.22GNGHH519 pKa = 4.96TAYY522 pKa = 10.14GRR524 pKa = 11.84SGGCIEE530 pKa = 4.49EE531 pKa = 4.2QVDD534 pKa = 3.8AYY536 pKa = 10.85LLEE539 pKa = 4.55GTTPAEE545 pKa = 4.45GARR548 pKa = 11.84CC549 pKa = 3.48
Molecular weight: 56.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8THF7|A0A4P8THF7_9MICO 1 4-dihydroxy-2-naphthoate octaprenyltransferase OS=Brachybacterium sp. SGAir0954 OX=2571029 GN=menA PE=3 SV=1
MM1 pKa = 7.28GSVVKK6 pKa = 10.44KK7 pKa = 9.48RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.32RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.28GSVVKK6 pKa = 10.44KK7 pKa = 9.48RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.32RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.79HH17 pKa = 5.67RR18 pKa = 11.84KK19 pKa = 8.31LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1012604 |
32 |
2236 |
342.9 |
36.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.721 ± 0.07 | 0.511 ± 0.01 |
6.137 ± 0.037 | 6.323 ± 0.052 |
2.612 ± 0.031 | 9.442 ± 0.036 |
2.194 ± 0.023 | 3.76 ± 0.031 |
1.619 ± 0.032 | 10.751 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.817 ± 0.018 | 1.475 ± 0.021 |
5.811 ± 0.035 | 2.737 ± 0.025 |
7.714 ± 0.057 | 5.422 ± 0.035 |
6.111 ± 0.038 | 8.677 ± 0.044 |
1.41 ± 0.02 | 1.757 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
