
Halobonum sp. NJ-3-1
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halobaculum
Average proteome isoelectric point is 4.94
Get precalculated fractions of proteins
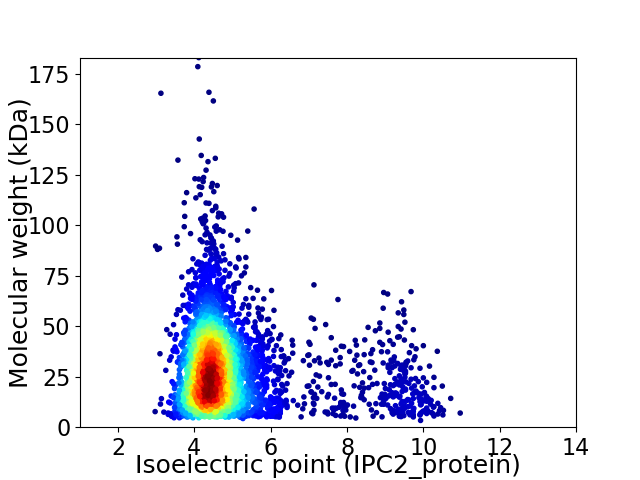
Virtual 2D-PAGE plot for 4124 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7D5LCI9|A0A7D5LCI9_9EURY F0F1 ATP synthase subunit C OS=Halobonum sp. NJ-3-1 OX=2743089 GN=HUG12_18085 PE=4 SV=1
MM1 pKa = 7.76TDD3 pKa = 3.41RR4 pKa = 11.84DD5 pKa = 4.16AAPDD9 pKa = 3.97LYY11 pKa = 11.06RR12 pKa = 11.84RR13 pKa = 11.84GLLKK17 pKa = 10.63AAGLAAGGAALPGVTSATPGRR38 pKa = 11.84EE39 pKa = 3.76PGAKK43 pKa = 9.13EE44 pKa = 4.4DD45 pKa = 3.84EE46 pKa = 4.55VLVGVSATADD56 pKa = 3.56DD57 pKa = 4.18VEE59 pKa = 4.78GAVARR64 pKa = 11.84HH65 pKa = 5.13VPGDD69 pKa = 3.65AEE71 pKa = 4.41VVHH74 pKa = 7.25SNEE77 pKa = 3.58TLRR80 pKa = 11.84YY81 pKa = 8.15VAVKK85 pKa = 9.83FPSGAADD92 pKa = 3.3VARR95 pKa = 11.84EE96 pKa = 3.98NFVDD100 pKa = 5.6AITKK104 pKa = 9.94KK105 pKa = 9.86EE106 pKa = 3.95HH107 pKa = 5.5VKK109 pKa = 10.5YY110 pKa = 10.71AEE112 pKa = 4.16PNATHH117 pKa = 7.27RR118 pKa = 11.84AFATPNDD125 pKa = 4.0PQFGDD130 pKa = 3.35QYY132 pKa = 11.78APQQVDD138 pKa = 5.32APDD141 pKa = 3.56AWDD144 pKa = 3.45VTYY147 pKa = 10.76GSSSVTVAVVDD158 pKa = 3.86TGAQYY163 pKa = 10.83DD164 pKa = 4.19HH165 pKa = 7.95PDD167 pKa = 3.45LADD170 pKa = 3.94NFADD174 pKa = 4.32DD175 pKa = 4.52PGYY178 pKa = 11.21DD179 pKa = 3.72FADD182 pKa = 4.97DD183 pKa = 5.58DD184 pKa = 4.75SDD186 pKa = 4.54PYY188 pKa = 11.05PDD190 pKa = 4.36APSDD194 pKa = 3.92EE195 pKa = 4.34YY196 pKa = 11.25HH197 pKa = 5.8GTHH200 pKa = 5.73VAGIAGAVTDD210 pKa = 4.34NGTGVAGVSDD220 pKa = 4.04SEE222 pKa = 4.81LINGRR227 pKa = 11.84ALDD230 pKa = 3.91EE231 pKa = 4.75SGSGSTADD239 pKa = 3.12IADD242 pKa = 4.7AIEE245 pKa = 3.73WAADD249 pKa = 3.67RR250 pKa = 11.84DD251 pKa = 3.91ADD253 pKa = 4.55VLNLSLGGGGYY264 pKa = 9.07TSTMKK269 pKa = 10.7NAVSYY274 pKa = 10.53AYY276 pKa = 10.55DD277 pKa = 3.28NGVFIAAAAGNDD289 pKa = 3.67GTNSVSYY296 pKa = 9.12PAAYY300 pKa = 9.87SEE302 pKa = 4.54CLAVSAVDD310 pKa = 3.85SNGDD314 pKa = 3.34LAYY317 pKa = 10.64FSTTGEE323 pKa = 4.32KK324 pKa = 10.55VEE326 pKa = 4.17LAAPGVDD333 pKa = 3.32VLSTTTEE340 pKa = 3.6ARR342 pKa = 11.84GSYY345 pKa = 7.52EE346 pKa = 4.12TLSGTSMATPVVAGSAGLALAQWDD370 pKa = 3.8VDD372 pKa = 3.58NATLRR377 pKa = 11.84SHH379 pKa = 7.11LKK381 pKa = 9.21DD382 pKa = 3.04TATDD386 pKa = 3.82LGLSSDD392 pKa = 4.03EE393 pKa = 4.18QGSGQVDD400 pKa = 3.57AGAAVATEE408 pKa = 4.41PGDD411 pKa = 3.72GGGGGGGGDD420 pKa = 3.51TTTGSVTDD428 pKa = 4.03SLSGYY433 pKa = 8.61WDD435 pKa = 3.41SDD437 pKa = 3.4CWSWAWEE444 pKa = 3.91YY445 pKa = 11.38GSPSKK450 pKa = 11.09VVIEE454 pKa = 4.39LDD456 pKa = 3.8GPSDD460 pKa = 4.4ADD462 pKa = 3.16FDD464 pKa = 4.96LYY466 pKa = 11.51ANEE469 pKa = 4.99GDD471 pKa = 4.19GTCPTTSSYY480 pKa = 11.37DD481 pKa = 3.52YY482 pKa = 10.58RR483 pKa = 11.84SWSTDD488 pKa = 2.93SQEE491 pKa = 4.57TITVDD496 pKa = 5.02DD497 pKa = 5.53PDD499 pKa = 3.88TSTDD503 pKa = 3.11LHH505 pKa = 7.46VLVDD509 pKa = 4.14SYY511 pKa = 11.59SGSGEE516 pKa = 3.89YY517 pKa = 10.14TLTITEE523 pKa = 4.43HH524 pKa = 5.87EE525 pKa = 4.27
MM1 pKa = 7.76TDD3 pKa = 3.41RR4 pKa = 11.84DD5 pKa = 4.16AAPDD9 pKa = 3.97LYY11 pKa = 11.06RR12 pKa = 11.84RR13 pKa = 11.84GLLKK17 pKa = 10.63AAGLAAGGAALPGVTSATPGRR38 pKa = 11.84EE39 pKa = 3.76PGAKK43 pKa = 9.13EE44 pKa = 4.4DD45 pKa = 3.84EE46 pKa = 4.55VLVGVSATADD56 pKa = 3.56DD57 pKa = 4.18VEE59 pKa = 4.78GAVARR64 pKa = 11.84HH65 pKa = 5.13VPGDD69 pKa = 3.65AEE71 pKa = 4.41VVHH74 pKa = 7.25SNEE77 pKa = 3.58TLRR80 pKa = 11.84YY81 pKa = 8.15VAVKK85 pKa = 9.83FPSGAADD92 pKa = 3.3VARR95 pKa = 11.84EE96 pKa = 3.98NFVDD100 pKa = 5.6AITKK104 pKa = 9.94KK105 pKa = 9.86EE106 pKa = 3.95HH107 pKa = 5.5VKK109 pKa = 10.5YY110 pKa = 10.71AEE112 pKa = 4.16PNATHH117 pKa = 7.27RR118 pKa = 11.84AFATPNDD125 pKa = 4.0PQFGDD130 pKa = 3.35QYY132 pKa = 11.78APQQVDD138 pKa = 5.32APDD141 pKa = 3.56AWDD144 pKa = 3.45VTYY147 pKa = 10.76GSSSVTVAVVDD158 pKa = 3.86TGAQYY163 pKa = 10.83DD164 pKa = 4.19HH165 pKa = 7.95PDD167 pKa = 3.45LADD170 pKa = 3.94NFADD174 pKa = 4.32DD175 pKa = 4.52PGYY178 pKa = 11.21DD179 pKa = 3.72FADD182 pKa = 4.97DD183 pKa = 5.58DD184 pKa = 4.75SDD186 pKa = 4.54PYY188 pKa = 11.05PDD190 pKa = 4.36APSDD194 pKa = 3.92EE195 pKa = 4.34YY196 pKa = 11.25HH197 pKa = 5.8GTHH200 pKa = 5.73VAGIAGAVTDD210 pKa = 4.34NGTGVAGVSDD220 pKa = 4.04SEE222 pKa = 4.81LINGRR227 pKa = 11.84ALDD230 pKa = 3.91EE231 pKa = 4.75SGSGSTADD239 pKa = 3.12IADD242 pKa = 4.7AIEE245 pKa = 3.73WAADD249 pKa = 3.67RR250 pKa = 11.84DD251 pKa = 3.91ADD253 pKa = 4.55VLNLSLGGGGYY264 pKa = 9.07TSTMKK269 pKa = 10.7NAVSYY274 pKa = 10.53AYY276 pKa = 10.55DD277 pKa = 3.28NGVFIAAAAGNDD289 pKa = 3.67GTNSVSYY296 pKa = 9.12PAAYY300 pKa = 9.87SEE302 pKa = 4.54CLAVSAVDD310 pKa = 3.85SNGDD314 pKa = 3.34LAYY317 pKa = 10.64FSTTGEE323 pKa = 4.32KK324 pKa = 10.55VEE326 pKa = 4.17LAAPGVDD333 pKa = 3.32VLSTTTEE340 pKa = 3.6ARR342 pKa = 11.84GSYY345 pKa = 7.52EE346 pKa = 4.12TLSGTSMATPVVAGSAGLALAQWDD370 pKa = 3.8VDD372 pKa = 3.58NATLRR377 pKa = 11.84SHH379 pKa = 7.11LKK381 pKa = 9.21DD382 pKa = 3.04TATDD386 pKa = 3.82LGLSSDD392 pKa = 4.03EE393 pKa = 4.18QGSGQVDD400 pKa = 3.57AGAAVATEE408 pKa = 4.41PGDD411 pKa = 3.72GGGGGGGGDD420 pKa = 3.51TTTGSVTDD428 pKa = 4.03SLSGYY433 pKa = 8.61WDD435 pKa = 3.41SDD437 pKa = 3.4CWSWAWEE444 pKa = 3.91YY445 pKa = 11.38GSPSKK450 pKa = 11.09VVIEE454 pKa = 4.39LDD456 pKa = 3.8GPSDD460 pKa = 4.4ADD462 pKa = 3.16FDD464 pKa = 4.96LYY466 pKa = 11.51ANEE469 pKa = 4.99GDD471 pKa = 4.19GTCPTTSSYY480 pKa = 11.37DD481 pKa = 3.52YY482 pKa = 10.58RR483 pKa = 11.84SWSTDD488 pKa = 2.93SQEE491 pKa = 4.57TITVDD496 pKa = 5.02DD497 pKa = 5.53PDD499 pKa = 3.88TSTDD503 pKa = 3.11LHH505 pKa = 7.46VLVDD509 pKa = 4.14SYY511 pKa = 11.59SGSGEE516 pKa = 3.89YY517 pKa = 10.14TLTITEE523 pKa = 4.43HH524 pKa = 5.87EE525 pKa = 4.27
Molecular weight: 53.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7D5LDS0|A0A7D5LDS0_9EURY UDP-ManNAc 6-dehydrogenase OS=Halobonum sp. NJ-3-1 OX=2743089 GN=HUG12_20275 PE=4 SV=1
MM1 pKa = 7.7ASFDD5 pKa = 3.5AAEE8 pKa = 3.99RR9 pKa = 11.84RR10 pKa = 11.84TLNKK14 pKa = 9.03MICMRR19 pKa = 11.84CNARR23 pKa = 11.84NSEE26 pKa = 3.99RR27 pKa = 11.84ADD29 pKa = 3.53RR30 pKa = 11.84CRR32 pKa = 11.84KK33 pKa = 9.5CGYY36 pKa = 9.92SRR38 pKa = 11.84LRR40 pKa = 11.84PKK42 pKa = 10.67AKK44 pKa = 10.13EE45 pKa = 3.68PRR47 pKa = 11.84TAA49 pKa = 3.84
MM1 pKa = 7.7ASFDD5 pKa = 3.5AAEE8 pKa = 3.99RR9 pKa = 11.84RR10 pKa = 11.84TLNKK14 pKa = 9.03MICMRR19 pKa = 11.84CNARR23 pKa = 11.84NSEE26 pKa = 3.99RR27 pKa = 11.84ADD29 pKa = 3.53RR30 pKa = 11.84CRR32 pKa = 11.84KK33 pKa = 9.5CGYY36 pKa = 9.92SRR38 pKa = 11.84LRR40 pKa = 11.84PKK42 pKa = 10.67AKK44 pKa = 10.13EE45 pKa = 3.68PRR47 pKa = 11.84TAA49 pKa = 3.84
Molecular weight: 5.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1167256 |
30 |
1656 |
283.0 |
30.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.474 ± 0.06 | 0.703 ± 0.015 |
8.268 ± 0.046 | 8.524 ± 0.057 |
3.309 ± 0.027 | 9.534 ± 0.035 |
2.002 ± 0.021 | 3.167 ± 0.036 |
1.56 ± 0.02 | 8.993 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.684 ± 0.018 | 2.177 ± 0.021 |
4.959 ± 0.031 | 1.94 ± 0.025 |
7.152 ± 0.043 | 5.171 ± 0.031 |
6.044 ± 0.034 | 9.626 ± 0.046 |
1.125 ± 0.015 | 2.587 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
