
Bovine parainfluenza 3 virus (BPIV-3)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Respirovirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
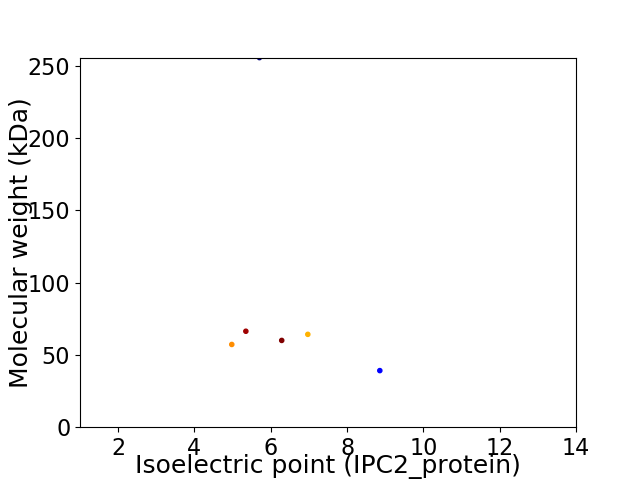
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6N3B3|A0A0F6N3B3_PI3B RNA-directed RNA polymerase L OS=Bovine parainfluenza 3 virus OX=11215 GN=L PE=3 SV=1
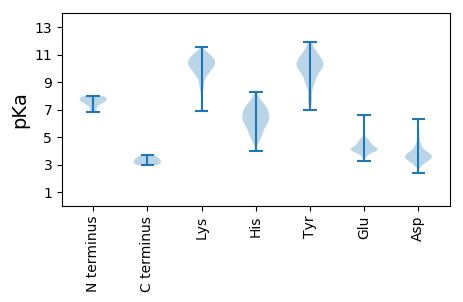
MM1 pKa = 7.66LSLFDD6 pKa = 3.55TFSARR11 pKa = 11.84RR12 pKa = 11.84QEE14 pKa = 4.8NITKK18 pKa = 10.01SAGGAVIPGQKK29 pKa = 8.52NTVSIFALGPSITDD43 pKa = 4.2DD44 pKa = 3.65NDD46 pKa = 3.31KK47 pKa = 8.61MTLALLFLSHH57 pKa = 7.05ALDD60 pKa = 3.55NEE62 pKa = 4.22KK63 pKa = 10.1QHH65 pKa = 5.8AQRR68 pKa = 11.84AGFLVSLLSMAYY80 pKa = 10.11ANPEE84 pKa = 4.09LYY86 pKa = 9.32LTANGSNADD95 pKa = 3.69VKK97 pKa = 10.62YY98 pKa = 10.03VIYY101 pKa = 9.65MIEE104 pKa = 4.29KK105 pKa = 9.95DD106 pKa = 3.63PGRR109 pKa = 11.84QKK111 pKa = 11.17YY112 pKa = 10.05GGLIIKK118 pKa = 7.97TRR120 pKa = 11.84EE121 pKa = 3.61MVYY124 pKa = 10.81EE125 pKa = 4.22KK126 pKa = 8.77TTDD129 pKa = 2.75WMFGSDD135 pKa = 5.39LEE137 pKa = 4.17YY138 pKa = 11.13DD139 pKa = 3.85QDD141 pKa = 4.46NMLQNGRR148 pKa = 11.84STSTIEE154 pKa = 4.64DD155 pKa = 3.65LVHH158 pKa = 5.73TFGYY162 pKa = 9.26PSCLGALIIQIWIILVKK179 pKa = 10.56AITSISGLRR188 pKa = 11.84KK189 pKa = 9.95GFFTRR194 pKa = 11.84LEE196 pKa = 4.02AFRR199 pKa = 11.84QDD201 pKa = 3.25GTVKK205 pKa = 10.6SSLVLSGDD213 pKa = 3.29AVEE216 pKa = 4.69QIGSIMRR223 pKa = 11.84SQQSLVTLMVEE234 pKa = 4.35TLITMNTGRR243 pKa = 11.84NDD245 pKa = 2.97LTTIEE250 pKa = 4.61KK251 pKa = 10.41NIQIVGNYY259 pKa = 8.94IRR261 pKa = 11.84DD262 pKa = 3.47AGLASFFNTIRR273 pKa = 11.84YY274 pKa = 9.22GIEE277 pKa = 3.27TRR279 pKa = 11.84MAALTLSTLRR289 pKa = 11.84PDD291 pKa = 3.44INRR294 pKa = 11.84LKK296 pKa = 11.05ALMEE300 pKa = 4.81LYY302 pKa = 10.37LSKK305 pKa = 10.93GPRR308 pKa = 11.84APFICILRR316 pKa = 11.84DD317 pKa = 3.48PVHH320 pKa = 7.27GEE322 pKa = 4.12FAPGNYY328 pKa = 7.59PALWSYY334 pKa = 11.87AMGVAVVQNKK344 pKa = 9.23AMQQYY349 pKa = 7.85VTGRR353 pKa = 11.84SYY355 pKa = 11.79LDD357 pKa = 2.92IEE359 pKa = 4.49MFQLGQAVARR369 pKa = 11.84DD370 pKa = 4.22AEE372 pKa = 4.51SQMSSILEE380 pKa = 4.26DD381 pKa = 3.33EE382 pKa = 4.8LGITQEE388 pKa = 4.54AKK390 pKa = 10.37QSLKK394 pKa = 10.79NHH396 pKa = 6.09MKK398 pKa = 10.42RR399 pKa = 11.84ISSSDD404 pKa = 3.21TAFQKK409 pKa = 9.39PTGGAAIEE417 pKa = 3.94MAIDD421 pKa = 3.76EE422 pKa = 4.62EE423 pKa = 4.96AEE425 pKa = 4.21QPDD428 pKa = 3.61SRR430 pKa = 11.84GDD432 pKa = 3.42QDD434 pKa = 3.78QSNGTQSSIIPYY446 pKa = 10.06AWADD450 pKa = 3.47DD451 pKa = 4.31TNNGTQTEE459 pKa = 4.69SATEE463 pKa = 3.69IDD465 pKa = 5.19SITTEE470 pKa = 3.58QRR472 pKa = 11.84NIRR475 pKa = 11.84DD476 pKa = 3.76RR477 pKa = 11.84LNKK480 pKa = 9.93RR481 pKa = 11.84LNEE484 pKa = 3.76KK485 pKa = 10.29RR486 pKa = 11.84KK487 pKa = 9.52QNEE490 pKa = 3.85LRR492 pKa = 11.84SDD494 pKa = 3.64DD495 pKa = 3.78TTDD498 pKa = 3.06NTNQTEE504 pKa = 3.99IDD506 pKa = 3.87DD507 pKa = 4.37LFSAFGSNN515 pKa = 3.34
MM1 pKa = 7.66LSLFDD6 pKa = 3.55TFSARR11 pKa = 11.84RR12 pKa = 11.84QEE14 pKa = 4.8NITKK18 pKa = 10.01SAGGAVIPGQKK29 pKa = 8.52NTVSIFALGPSITDD43 pKa = 4.2DD44 pKa = 3.65NDD46 pKa = 3.31KK47 pKa = 8.61MTLALLFLSHH57 pKa = 7.05ALDD60 pKa = 3.55NEE62 pKa = 4.22KK63 pKa = 10.1QHH65 pKa = 5.8AQRR68 pKa = 11.84AGFLVSLLSMAYY80 pKa = 10.11ANPEE84 pKa = 4.09LYY86 pKa = 9.32LTANGSNADD95 pKa = 3.69VKK97 pKa = 10.62YY98 pKa = 10.03VIYY101 pKa = 9.65MIEE104 pKa = 4.29KK105 pKa = 9.95DD106 pKa = 3.63PGRR109 pKa = 11.84QKK111 pKa = 11.17YY112 pKa = 10.05GGLIIKK118 pKa = 7.97TRR120 pKa = 11.84EE121 pKa = 3.61MVYY124 pKa = 10.81EE125 pKa = 4.22KK126 pKa = 8.77TTDD129 pKa = 2.75WMFGSDD135 pKa = 5.39LEE137 pKa = 4.17YY138 pKa = 11.13DD139 pKa = 3.85QDD141 pKa = 4.46NMLQNGRR148 pKa = 11.84STSTIEE154 pKa = 4.64DD155 pKa = 3.65LVHH158 pKa = 5.73TFGYY162 pKa = 9.26PSCLGALIIQIWIILVKK179 pKa = 10.56AITSISGLRR188 pKa = 11.84KK189 pKa = 9.95GFFTRR194 pKa = 11.84LEE196 pKa = 4.02AFRR199 pKa = 11.84QDD201 pKa = 3.25GTVKK205 pKa = 10.6SSLVLSGDD213 pKa = 3.29AVEE216 pKa = 4.69QIGSIMRR223 pKa = 11.84SQQSLVTLMVEE234 pKa = 4.35TLITMNTGRR243 pKa = 11.84NDD245 pKa = 2.97LTTIEE250 pKa = 4.61KK251 pKa = 10.41NIQIVGNYY259 pKa = 8.94IRR261 pKa = 11.84DD262 pKa = 3.47AGLASFFNTIRR273 pKa = 11.84YY274 pKa = 9.22GIEE277 pKa = 3.27TRR279 pKa = 11.84MAALTLSTLRR289 pKa = 11.84PDD291 pKa = 3.44INRR294 pKa = 11.84LKK296 pKa = 11.05ALMEE300 pKa = 4.81LYY302 pKa = 10.37LSKK305 pKa = 10.93GPRR308 pKa = 11.84APFICILRR316 pKa = 11.84DD317 pKa = 3.48PVHH320 pKa = 7.27GEE322 pKa = 4.12FAPGNYY328 pKa = 7.59PALWSYY334 pKa = 11.87AMGVAVVQNKK344 pKa = 9.23AMQQYY349 pKa = 7.85VTGRR353 pKa = 11.84SYY355 pKa = 11.79LDD357 pKa = 2.92IEE359 pKa = 4.49MFQLGQAVARR369 pKa = 11.84DD370 pKa = 4.22AEE372 pKa = 4.51SQMSSILEE380 pKa = 4.26DD381 pKa = 3.33EE382 pKa = 4.8LGITQEE388 pKa = 4.54AKK390 pKa = 10.37QSLKK394 pKa = 10.79NHH396 pKa = 6.09MKK398 pKa = 10.42RR399 pKa = 11.84ISSSDD404 pKa = 3.21TAFQKK409 pKa = 9.39PTGGAAIEE417 pKa = 3.94MAIDD421 pKa = 3.76EE422 pKa = 4.62EE423 pKa = 4.96AEE425 pKa = 4.21QPDD428 pKa = 3.61SRR430 pKa = 11.84GDD432 pKa = 3.42QDD434 pKa = 3.78QSNGTQSSIIPYY446 pKa = 10.06AWADD450 pKa = 3.47DD451 pKa = 4.31TNNGTQTEE459 pKa = 4.69SATEE463 pKa = 3.69IDD465 pKa = 5.19SITTEE470 pKa = 3.58QRR472 pKa = 11.84NIRR475 pKa = 11.84DD476 pKa = 3.76RR477 pKa = 11.84LNKK480 pKa = 9.93RR481 pKa = 11.84LNEE484 pKa = 3.76KK485 pKa = 10.29RR486 pKa = 11.84KK487 pKa = 9.52QNEE490 pKa = 3.85LRR492 pKa = 11.84SDD494 pKa = 3.64DD495 pKa = 3.78TTDD498 pKa = 3.06NTNQTEE504 pKa = 3.99IDD506 pKa = 3.87DD507 pKa = 4.37LFSAFGSNN515 pKa = 3.34
Molecular weight: 57.27 kDa
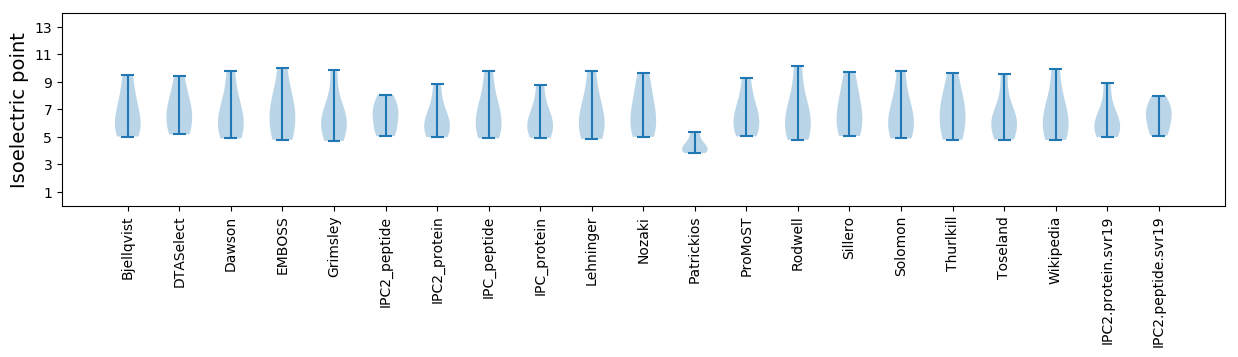
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6N3J2|A0A0F6N3J2_PI3B Fusion glycoprotein F0 OS=Bovine parainfluenza 3 virus OX=11215 GN=F PE=3 SV=1
MM1 pKa = 7.57SIKK4 pKa = 10.59NSAIYY9 pKa = 8.33TFPDD13 pKa = 3.74SSFLDD18 pKa = 3.57NGNIEE23 pKa = 4.19PLPLKK28 pKa = 10.86VNEE31 pKa = 3.85QRR33 pKa = 11.84KK34 pKa = 8.51AVPHH38 pKa = 5.6IRR40 pKa = 11.84VVKK43 pKa = 10.55IGDD46 pKa = 3.75PPKK49 pKa = 10.55HH50 pKa = 5.98GSRR53 pKa = 11.84YY54 pKa = 10.04LDD56 pKa = 3.28VFLLGFFEE64 pKa = 4.52MEE66 pKa = 3.99RR67 pKa = 11.84SKK69 pKa = 11.36DD70 pKa = 3.45RR71 pKa = 11.84YY72 pKa = 10.76GSISDD77 pKa = 4.9LDD79 pKa = 4.49DD80 pKa = 4.59DD81 pKa = 4.99PSYY84 pKa = 10.83KK85 pKa = 10.39VCGSGSLPLGLAKK98 pKa = 9.91YY99 pKa = 8.79TGGDD103 pKa = 3.49QEE105 pKa = 4.96LLQAAIKK112 pKa = 10.62LDD114 pKa = 3.28IEE116 pKa = 4.2VRR118 pKa = 11.84RR119 pKa = 11.84TVKK122 pKa = 8.8ATEE125 pKa = 4.15MIVYY129 pKa = 7.68TVQNIKK135 pKa = 10.55PEE137 pKa = 4.48LYY139 pKa = 8.64PWSGRR144 pKa = 11.84LRR146 pKa = 11.84KK147 pKa = 10.13GMLFDD152 pKa = 4.35ANKK155 pKa = 10.02VALAPQCLPLDD166 pKa = 3.73RR167 pKa = 11.84GIKK170 pKa = 9.81FRR172 pKa = 11.84VIFVNCTAIGSITLFKK188 pKa = 10.31IPKK191 pKa = 9.93SMALLSLPNTISINLQVHH209 pKa = 6.55IKK211 pKa = 9.71TGIQTDD217 pKa = 3.63SKK219 pKa = 11.29GVVQILDD226 pKa = 3.61EE227 pKa = 4.79KK228 pKa = 11.28GEE230 pKa = 3.98KK231 pKa = 9.96SLNFMVHH238 pKa = 6.48LGLIKK243 pKa = 10.4RR244 pKa = 11.84KK245 pKa = 8.81IGRR248 pKa = 11.84MYY250 pKa = 10.53SVEE253 pKa = 3.94YY254 pKa = 10.17CKK256 pKa = 10.57QKK258 pKa = 9.97IEE260 pKa = 4.15KK261 pKa = 8.94MRR263 pKa = 11.84LLFSLGLVGGISFHH277 pKa = 6.95VNATGSISKK286 pKa = 8.63TLASQLAFKK295 pKa = 10.94RR296 pKa = 11.84EE297 pKa = 3.5ICYY300 pKa = 10.23PLMDD304 pKa = 5.94LNPHH308 pKa = 6.39LNSVIWASSIEE319 pKa = 4.07ITRR322 pKa = 11.84VDD324 pKa = 5.07AILQPSLPGEE334 pKa = 3.7FRR336 pKa = 11.84YY337 pKa = 10.34YY338 pKa = 10.39PNIIAKK344 pKa = 9.88GVGKK348 pKa = 10.41IKK350 pKa = 10.5QQ351 pKa = 3.19
MM1 pKa = 7.57SIKK4 pKa = 10.59NSAIYY9 pKa = 8.33TFPDD13 pKa = 3.74SSFLDD18 pKa = 3.57NGNIEE23 pKa = 4.19PLPLKK28 pKa = 10.86VNEE31 pKa = 3.85QRR33 pKa = 11.84KK34 pKa = 8.51AVPHH38 pKa = 5.6IRR40 pKa = 11.84VVKK43 pKa = 10.55IGDD46 pKa = 3.75PPKK49 pKa = 10.55HH50 pKa = 5.98GSRR53 pKa = 11.84YY54 pKa = 10.04LDD56 pKa = 3.28VFLLGFFEE64 pKa = 4.52MEE66 pKa = 3.99RR67 pKa = 11.84SKK69 pKa = 11.36DD70 pKa = 3.45RR71 pKa = 11.84YY72 pKa = 10.76GSISDD77 pKa = 4.9LDD79 pKa = 4.49DD80 pKa = 4.59DD81 pKa = 4.99PSYY84 pKa = 10.83KK85 pKa = 10.39VCGSGSLPLGLAKK98 pKa = 9.91YY99 pKa = 8.79TGGDD103 pKa = 3.49QEE105 pKa = 4.96LLQAAIKK112 pKa = 10.62LDD114 pKa = 3.28IEE116 pKa = 4.2VRR118 pKa = 11.84RR119 pKa = 11.84TVKK122 pKa = 8.8ATEE125 pKa = 4.15MIVYY129 pKa = 7.68TVQNIKK135 pKa = 10.55PEE137 pKa = 4.48LYY139 pKa = 8.64PWSGRR144 pKa = 11.84LRR146 pKa = 11.84KK147 pKa = 10.13GMLFDD152 pKa = 4.35ANKK155 pKa = 10.02VALAPQCLPLDD166 pKa = 3.73RR167 pKa = 11.84GIKK170 pKa = 9.81FRR172 pKa = 11.84VIFVNCTAIGSITLFKK188 pKa = 10.31IPKK191 pKa = 9.93SMALLSLPNTISINLQVHH209 pKa = 6.55IKK211 pKa = 9.71TGIQTDD217 pKa = 3.63SKK219 pKa = 11.29GVVQILDD226 pKa = 3.61EE227 pKa = 4.79KK228 pKa = 11.28GEE230 pKa = 3.98KK231 pKa = 9.96SLNFMVHH238 pKa = 6.48LGLIKK243 pKa = 10.4RR244 pKa = 11.84KK245 pKa = 8.81IGRR248 pKa = 11.84MYY250 pKa = 10.53SVEE253 pKa = 3.94YY254 pKa = 10.17CKK256 pKa = 10.57QKK258 pKa = 9.97IEE260 pKa = 4.15KK261 pKa = 8.94MRR263 pKa = 11.84LLFSLGLVGGISFHH277 pKa = 6.95VNATGSISKK286 pKa = 8.63TLASQLAFKK295 pKa = 10.94RR296 pKa = 11.84EE297 pKa = 3.5ICYY300 pKa = 10.23PLMDD304 pKa = 5.94LNPHH308 pKa = 6.39LNSVIWASSIEE319 pKa = 4.07ITRR322 pKa = 11.84VDD324 pKa = 5.07AILQPSLPGEE334 pKa = 3.7FRR336 pKa = 11.84YY337 pKa = 10.34YY338 pKa = 10.39PNIIAKK344 pKa = 9.88GVGKK348 pKa = 10.41IKK350 pKa = 10.5QQ351 pKa = 3.19
Molecular weight: 39.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4807 |
351 |
2233 |
801.2 |
90.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.722 ± 0.473 | 1.435 ± 0.277 |
6.137 ± 0.276 | 5.804 ± 0.694 |
3.016 ± 0.369 | 5.596 ± 0.47 |
1.685 ± 0.224 | 9.341 ± 0.843 |
6.532 ± 0.449 | 9.673 ± 0.702 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.274 | 6.511 ± 0.402 |
3.828 ± 0.294 | 3.828 ± 0.543 |
4.681 ± 0.38 | 8.134 ± 0.329 |
6.782 ± 0.58 | 4.91 ± 0.388 |
1.144 ± 0.213 | 3.828 ± 0.451 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
