
Hubei tombus-like virus 19
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.69
Get precalculated fractions of proteins
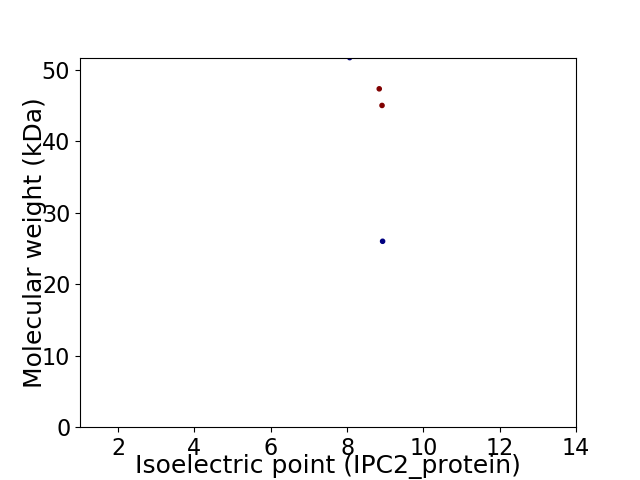
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGU0|A0A1L3KGU0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 19 OX=1923265 PE=4 SV=1
MM1 pKa = 7.71EE2 pKa = 5.09NLSRR6 pKa = 11.84PSSLSCGKK14 pKa = 8.68FTMEE18 pKa = 4.07KK19 pKa = 10.25SKK21 pKa = 9.49TRR23 pKa = 11.84SPRR26 pKa = 11.84STNLRR31 pKa = 11.84EE32 pKa = 4.21SNEE35 pKa = 4.15KK36 pKa = 10.21IVTNVLLARR45 pKa = 11.84LSEE48 pKa = 3.96QLEE51 pKa = 4.6TINRR55 pKa = 11.84RR56 pKa = 11.84ISLIEE61 pKa = 4.63DD62 pKa = 3.31SLQEE66 pKa = 4.25SNRR69 pKa = 11.84SQLQLILSISRR80 pKa = 11.84LEE82 pKa = 4.06KK83 pKa = 9.93EE84 pKa = 4.62CRR86 pKa = 11.84QGVPGANSPTEE97 pKa = 4.03SRR99 pKa = 11.84AKK101 pKa = 10.29SSKK104 pKa = 10.26KK105 pKa = 10.48LPSCRR110 pKa = 11.84TNPTIPPFRR119 pKa = 11.84SRR121 pKa = 11.84QEE123 pKa = 3.98SFSSIDD129 pKa = 3.5SLEE132 pKa = 4.0SMYY135 pKa = 10.49PGPPQRR141 pKa = 11.84HH142 pKa = 6.22LGRR145 pKa = 11.84PAMNEE150 pKa = 3.51ALIRR154 pKa = 11.84EE155 pKa = 4.71LNTTLHH161 pKa = 6.67ASCGGQTASSSHH173 pKa = 7.12PDD175 pKa = 3.21SEE177 pKa = 4.78GCGIVSPGDD186 pKa = 3.54NNNEE190 pKa = 4.19SVPTKK195 pKa = 10.01PSPLTSQSACISGPQIEE212 pKa = 5.21PKK214 pKa = 10.14RR215 pKa = 11.84QAPQPPVEE223 pKa = 4.38EE224 pKa = 4.39VVPPAQPPAVNGAQPNVTEE243 pKa = 4.43EE244 pKa = 4.09VAKK247 pKa = 8.95TVCAQPKK254 pKa = 8.57ATMRR258 pKa = 11.84PKK260 pKa = 9.51LTHH263 pKa = 6.62PEE265 pKa = 3.62EE266 pKa = 4.07TADD269 pKa = 4.67YY270 pKa = 10.28IISTNDD276 pKa = 2.75SRR278 pKa = 11.84LVKK281 pKa = 9.95EE282 pKa = 4.37ALGVTPQIVVSSSVDD297 pKa = 3.04QTLRR301 pKa = 11.84WRR303 pKa = 11.84ATKK306 pKa = 9.91IDD308 pKa = 3.74GEE310 pKa = 4.6VVLRR314 pKa = 11.84SEE316 pKa = 4.51PVPVIRR322 pKa = 11.84SDD324 pKa = 3.06AGYY327 pKa = 10.03FSNSGYY333 pKa = 9.94YY334 pKa = 10.35AEE336 pKa = 5.98DD337 pKa = 3.25EE338 pKa = 4.25TRR340 pKa = 11.84PAKK343 pKa = 9.69TWVIGPAAPVRR354 pKa = 11.84LIAWVDD360 pKa = 3.5HH361 pKa = 6.12EE362 pKa = 4.75MLAHH366 pKa = 5.34MRR368 pKa = 11.84RR369 pKa = 11.84YY370 pKa = 9.1IMKK373 pKa = 10.29VGLTQSSYY381 pKa = 11.36HH382 pKa = 6.37KK383 pKa = 10.59LVAQGEE389 pKa = 4.48LFLKK393 pKa = 10.55NFRR396 pKa = 11.84TDD398 pKa = 2.97LTDD401 pKa = 2.86PHH403 pKa = 6.66YY404 pKa = 11.3VNRR407 pKa = 11.84ILDD410 pKa = 3.69STACVAMVPTQNEE423 pKa = 4.22VVNITKK429 pKa = 10.49LSSGLVFNAINTVSDD444 pKa = 3.95FRR446 pKa = 11.84RR447 pKa = 11.84LGLVPRR453 pKa = 11.84KK454 pKa = 9.32VWCGLFNGSPYY465 pKa = 10.64KK466 pKa = 10.74LPSATT471 pKa = 3.73
MM1 pKa = 7.71EE2 pKa = 5.09NLSRR6 pKa = 11.84PSSLSCGKK14 pKa = 8.68FTMEE18 pKa = 4.07KK19 pKa = 10.25SKK21 pKa = 9.49TRR23 pKa = 11.84SPRR26 pKa = 11.84STNLRR31 pKa = 11.84EE32 pKa = 4.21SNEE35 pKa = 4.15KK36 pKa = 10.21IVTNVLLARR45 pKa = 11.84LSEE48 pKa = 3.96QLEE51 pKa = 4.6TINRR55 pKa = 11.84RR56 pKa = 11.84ISLIEE61 pKa = 4.63DD62 pKa = 3.31SLQEE66 pKa = 4.25SNRR69 pKa = 11.84SQLQLILSISRR80 pKa = 11.84LEE82 pKa = 4.06KK83 pKa = 9.93EE84 pKa = 4.62CRR86 pKa = 11.84QGVPGANSPTEE97 pKa = 4.03SRR99 pKa = 11.84AKK101 pKa = 10.29SSKK104 pKa = 10.26KK105 pKa = 10.48LPSCRR110 pKa = 11.84TNPTIPPFRR119 pKa = 11.84SRR121 pKa = 11.84QEE123 pKa = 3.98SFSSIDD129 pKa = 3.5SLEE132 pKa = 4.0SMYY135 pKa = 10.49PGPPQRR141 pKa = 11.84HH142 pKa = 6.22LGRR145 pKa = 11.84PAMNEE150 pKa = 3.51ALIRR154 pKa = 11.84EE155 pKa = 4.71LNTTLHH161 pKa = 6.67ASCGGQTASSSHH173 pKa = 7.12PDD175 pKa = 3.21SEE177 pKa = 4.78GCGIVSPGDD186 pKa = 3.54NNNEE190 pKa = 4.19SVPTKK195 pKa = 10.01PSPLTSQSACISGPQIEE212 pKa = 5.21PKK214 pKa = 10.14RR215 pKa = 11.84QAPQPPVEE223 pKa = 4.38EE224 pKa = 4.39VVPPAQPPAVNGAQPNVTEE243 pKa = 4.43EE244 pKa = 4.09VAKK247 pKa = 8.95TVCAQPKK254 pKa = 8.57ATMRR258 pKa = 11.84PKK260 pKa = 9.51LTHH263 pKa = 6.62PEE265 pKa = 3.62EE266 pKa = 4.07TADD269 pKa = 4.67YY270 pKa = 10.28IISTNDD276 pKa = 2.75SRR278 pKa = 11.84LVKK281 pKa = 9.95EE282 pKa = 4.37ALGVTPQIVVSSSVDD297 pKa = 3.04QTLRR301 pKa = 11.84WRR303 pKa = 11.84ATKK306 pKa = 9.91IDD308 pKa = 3.74GEE310 pKa = 4.6VVLRR314 pKa = 11.84SEE316 pKa = 4.51PVPVIRR322 pKa = 11.84SDD324 pKa = 3.06AGYY327 pKa = 10.03FSNSGYY333 pKa = 9.94YY334 pKa = 10.35AEE336 pKa = 5.98DD337 pKa = 3.25EE338 pKa = 4.25TRR340 pKa = 11.84PAKK343 pKa = 9.69TWVIGPAAPVRR354 pKa = 11.84LIAWVDD360 pKa = 3.5HH361 pKa = 6.12EE362 pKa = 4.75MLAHH366 pKa = 5.34MRR368 pKa = 11.84RR369 pKa = 11.84YY370 pKa = 9.1IMKK373 pKa = 10.29VGLTQSSYY381 pKa = 11.36HH382 pKa = 6.37KK383 pKa = 10.59LVAQGEE389 pKa = 4.48LFLKK393 pKa = 10.55NFRR396 pKa = 11.84TDD398 pKa = 2.97LTDD401 pKa = 2.86PHH403 pKa = 6.66YY404 pKa = 11.3VNRR407 pKa = 11.84ILDD410 pKa = 3.69STACVAMVPTQNEE423 pKa = 4.22VVNITKK429 pKa = 10.49LSSGLVFNAINTVSDD444 pKa = 3.95FRR446 pKa = 11.84RR447 pKa = 11.84LGLVPRR453 pKa = 11.84KK454 pKa = 9.32VWCGLFNGSPYY465 pKa = 10.64KK466 pKa = 10.74LPSATT471 pKa = 3.73
Molecular weight: 51.74 kDa
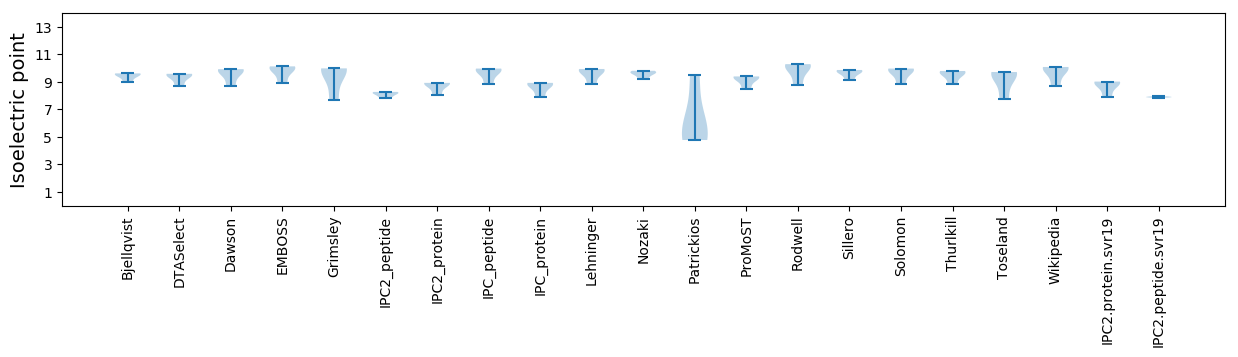
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGM9|A0A1L3KGM9_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 19 OX=1923265 PE=4 SV=1
MM1 pKa = 7.31QMPKK5 pKa = 10.09PKK7 pKa = 10.08SKK9 pKa = 9.11KK10 pKa = 6.28TLRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 8.82QPRR18 pKa = 11.84PRR20 pKa = 11.84QQRR23 pKa = 11.84PLAPMNVGARR33 pKa = 11.84LRR35 pKa = 11.84TAQSTRR41 pKa = 11.84KK42 pKa = 9.39KK43 pKa = 10.17PEE45 pKa = 3.71VVTDD49 pKa = 4.2LLLSVDD55 pKa = 3.83IPKK58 pKa = 8.82DD59 pKa = 3.62TPRR62 pKa = 11.84GTILLDD68 pKa = 3.05ISLNPTITKK77 pKa = 10.35RR78 pKa = 11.84LANLSKK84 pKa = 11.13AFVKK88 pKa = 10.02WKK90 pKa = 7.21MTNCEE95 pKa = 4.03IQVQTQVPATMGGGLVGGYY114 pKa = 9.92VPDD117 pKa = 4.05PSIRR121 pKa = 11.84LSDD124 pKa = 4.74GIQVLEE130 pKa = 4.54SLTATNQAKK139 pKa = 9.64IVKK142 pKa = 7.79TWEE145 pKa = 3.99SFTVKK150 pKa = 10.08IPNSKK155 pKa = 9.88EE156 pKa = 3.79LFVVRR161 pKa = 11.84GDD163 pKa = 4.13DD164 pKa = 4.64SRR166 pKa = 11.84LSDD169 pKa = 3.19AGRR172 pKa = 11.84FYY174 pKa = 11.28LVVDD178 pKa = 5.11GPPTDD183 pKa = 3.4NSSIRR188 pKa = 11.84VQIRR192 pKa = 11.84WSVQLLVPFAQDD204 pKa = 2.75EE205 pKa = 4.41RR206 pKa = 11.84NILDD210 pKa = 4.07SLTVTCSALGIQGEE224 pKa = 4.69GVLEE228 pKa = 4.3GSRR231 pKa = 11.84NCTVHH236 pKa = 6.05NWTPQHH242 pKa = 6.09GFAPPFDD249 pKa = 4.01KK250 pKa = 11.15TPDD253 pKa = 3.59LDD255 pKa = 4.36LSHH258 pKa = 6.84GLHH261 pKa = 6.51GLPNFSSAGPNIRR274 pKa = 11.84YY275 pKa = 9.04YY276 pKa = 10.94RR277 pKa = 11.84LDD279 pKa = 3.44QPAIAHH285 pKa = 6.47PAALFLAVFNQRR297 pKa = 11.84MLNASLPDD305 pKa = 3.09SWRR308 pKa = 11.84GVFLDD313 pKa = 3.55MPNYY317 pKa = 9.19PEE319 pKa = 4.96PDD321 pKa = 2.98ISVAPSIMLVQHH333 pKa = 5.66STLTPVLQEE342 pKa = 4.25EE343 pKa = 4.81YY344 pKa = 10.67LGNQEE349 pKa = 3.96GSFLVTVHH357 pKa = 6.84RR358 pKa = 11.84FLSATRR364 pKa = 11.84QLQCSKK370 pKa = 9.82EE371 pKa = 3.88YY372 pKa = 11.07RR373 pKa = 11.84MAQPNPLLTRR383 pKa = 11.84SLPQEE388 pKa = 3.91NVAMVLSRR396 pKa = 11.84KK397 pKa = 8.6LAQMSLVNN405 pKa = 4.04
MM1 pKa = 7.31QMPKK5 pKa = 10.09PKK7 pKa = 10.08SKK9 pKa = 9.11KK10 pKa = 6.28TLRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 8.82QPRR18 pKa = 11.84PRR20 pKa = 11.84QQRR23 pKa = 11.84PLAPMNVGARR33 pKa = 11.84LRR35 pKa = 11.84TAQSTRR41 pKa = 11.84KK42 pKa = 9.39KK43 pKa = 10.17PEE45 pKa = 3.71VVTDD49 pKa = 4.2LLLSVDD55 pKa = 3.83IPKK58 pKa = 8.82DD59 pKa = 3.62TPRR62 pKa = 11.84GTILLDD68 pKa = 3.05ISLNPTITKK77 pKa = 10.35RR78 pKa = 11.84LANLSKK84 pKa = 11.13AFVKK88 pKa = 10.02WKK90 pKa = 7.21MTNCEE95 pKa = 4.03IQVQTQVPATMGGGLVGGYY114 pKa = 9.92VPDD117 pKa = 4.05PSIRR121 pKa = 11.84LSDD124 pKa = 4.74GIQVLEE130 pKa = 4.54SLTATNQAKK139 pKa = 9.64IVKK142 pKa = 7.79TWEE145 pKa = 3.99SFTVKK150 pKa = 10.08IPNSKK155 pKa = 9.88EE156 pKa = 3.79LFVVRR161 pKa = 11.84GDD163 pKa = 4.13DD164 pKa = 4.64SRR166 pKa = 11.84LSDD169 pKa = 3.19AGRR172 pKa = 11.84FYY174 pKa = 11.28LVVDD178 pKa = 5.11GPPTDD183 pKa = 3.4NSSIRR188 pKa = 11.84VQIRR192 pKa = 11.84WSVQLLVPFAQDD204 pKa = 2.75EE205 pKa = 4.41RR206 pKa = 11.84NILDD210 pKa = 4.07SLTVTCSALGIQGEE224 pKa = 4.69GVLEE228 pKa = 4.3GSRR231 pKa = 11.84NCTVHH236 pKa = 6.05NWTPQHH242 pKa = 6.09GFAPPFDD249 pKa = 4.01KK250 pKa = 11.15TPDD253 pKa = 3.59LDD255 pKa = 4.36LSHH258 pKa = 6.84GLHH261 pKa = 6.51GLPNFSSAGPNIRR274 pKa = 11.84YY275 pKa = 9.04YY276 pKa = 10.94RR277 pKa = 11.84LDD279 pKa = 3.44QPAIAHH285 pKa = 6.47PAALFLAVFNQRR297 pKa = 11.84MLNASLPDD305 pKa = 3.09SWRR308 pKa = 11.84GVFLDD313 pKa = 3.55MPNYY317 pKa = 9.19PEE319 pKa = 4.96PDD321 pKa = 2.98ISVAPSIMLVQHH333 pKa = 5.66STLTPVLQEE342 pKa = 4.25EE343 pKa = 4.81YY344 pKa = 10.67LGNQEE349 pKa = 3.96GSFLVTVHH357 pKa = 6.84RR358 pKa = 11.84FLSATRR364 pKa = 11.84QLQCSKK370 pKa = 9.82EE371 pKa = 3.88YY372 pKa = 11.07RR373 pKa = 11.84MAQPNPLLTRR383 pKa = 11.84SLPQEE388 pKa = 3.91NVAMVLSRR396 pKa = 11.84KK397 pKa = 8.6LAQMSLVNN405 pKa = 4.04
Molecular weight: 45.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1516 |
228 |
471 |
379.0 |
42.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.728 ± 0.346 | 1.451 ± 0.184 |
4.485 ± 0.472 | 5.409 ± 0.654 |
3.826 ± 0.859 | 5.607 ± 0.309 |
1.847 ± 0.096 | 4.485 ± 0.267 |
5.739 ± 0.686 | 8.839 ± 0.857 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.441 ± 0.509 | 4.222 ± 0.426 |
7.058 ± 0.92 | 4.551 ± 0.553 |
7.19 ± 0.348 | 8.377 ± 1.174 |
6.069 ± 0.439 | 7.52 ± 0.173 |
1.715 ± 0.43 | 2.441 ± 0.382 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
