
Tomato necrotic streak virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
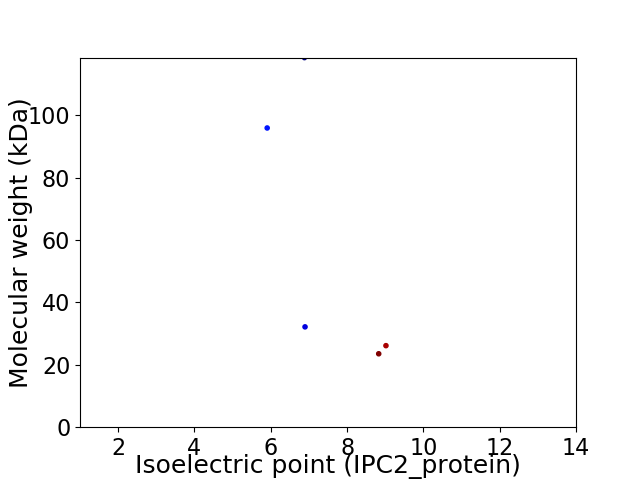
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140GPI7|A0A140GPI7_9BROM Putative silencing suppressor OS=Tomato necrotic streak virus OX=1637492 GN=2b PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 6.42IITNVLDD9 pKa = 3.87LVSTVSLRR17 pKa = 11.84TSLQVHH23 pKa = 6.7ASFDD27 pKa = 3.89LPLSIEE33 pKa = 3.85QSFMIRR39 pKa = 11.84LWLVRR44 pKa = 11.84DD45 pKa = 3.79VVIPTMKK52 pKa = 10.43VYY54 pKa = 10.24WDD56 pKa = 3.54ADD58 pKa = 3.44ALLGRR63 pKa = 11.84LEE65 pKa = 4.69TVFSTDD71 pKa = 3.18PEE73 pKa = 4.22PVVMRR78 pKa = 11.84NSTFHH83 pKa = 7.44DD84 pKa = 3.55IVPEE88 pKa = 3.93YY89 pKa = 10.58VDD91 pKa = 3.68EE92 pKa = 4.87FGEE95 pKa = 4.53VPEE98 pKa = 4.18EE99 pKa = 4.43AYY101 pKa = 10.72LSPEE105 pKa = 3.8EE106 pKa = 4.26RR107 pKa = 11.84EE108 pKa = 4.42EE109 pKa = 3.66IHH111 pKa = 6.87KK112 pKa = 9.54ATFLKK117 pKa = 10.33HH118 pKa = 5.15EE119 pKa = 4.29HH120 pKa = 6.52WGSEE124 pKa = 4.02SSGSSFLDD132 pKa = 4.26EE133 pKa = 4.74IEE135 pKa = 4.36CLYY138 pKa = 11.3DD139 pKa = 3.37EE140 pKa = 4.47TVKK143 pKa = 11.12DD144 pKa = 4.68DD145 pKa = 3.88VMQDD149 pKa = 2.74VCTDD153 pKa = 3.26MPKK156 pKa = 10.4DD157 pKa = 3.93YY158 pKa = 11.12EE159 pKa = 4.43IMWNDD164 pKa = 3.55GAVDD168 pKa = 3.98AVWSSRR174 pKa = 11.84FEE176 pKa = 4.02VEE178 pKa = 4.62EE179 pKa = 3.94PPKK182 pKa = 10.62PKK184 pKa = 9.93FRR186 pKa = 11.84PQMTDD191 pKa = 2.89KK192 pKa = 11.21VCDD195 pKa = 3.48PVIIQDD201 pKa = 4.7AINDD205 pKa = 3.88IFPFHH210 pKa = 7.22QDD212 pKa = 2.3MDD214 pKa = 3.76DD215 pKa = 3.76TYY217 pKa = 10.48FQTWVEE223 pKa = 4.15TQDD226 pKa = 3.07ISLEE230 pKa = 4.18VSKK233 pKa = 10.96CWLDD237 pKa = 3.26ASNFKK242 pKa = 10.96DD243 pKa = 3.78FTKK246 pKa = 10.69GQQTYY251 pKa = 10.4AVPTYY256 pKa = 10.83QSGATSKK263 pKa = 10.53RR264 pKa = 11.84VNTQRR269 pKa = 11.84EE270 pKa = 3.87TLLAVKK276 pKa = 9.35KK277 pKa = 10.43RR278 pKa = 11.84NMNIPEE284 pKa = 4.13LQATFDD290 pKa = 4.13LDD292 pKa = 4.62AEE294 pKa = 4.44VATCFKK300 pKa = 10.54RR301 pKa = 11.84FCTHH305 pKa = 5.53VVDD308 pKa = 4.49IPRR311 pKa = 11.84LKK313 pKa = 10.43RR314 pKa = 11.84LPPMMGTEE322 pKa = 4.05TEE324 pKa = 4.2FFHH327 pKa = 7.94RR328 pKa = 11.84LFGWYY333 pKa = 9.7NPPLSEE339 pKa = 4.06YY340 pKa = 10.36RR341 pKa = 11.84GPLTLQSLDD350 pKa = 3.65KK351 pKa = 10.5YY352 pKa = 8.34MHH354 pKa = 5.5MVKK357 pKa = 9.98TIVKK361 pKa = 8.75PVEE364 pKa = 4.43DD365 pKa = 3.85NSLKK369 pKa = 10.61YY370 pKa = 10.06EE371 pKa = 4.23RR372 pKa = 11.84PLCATITLSKK382 pKa = 10.3KK383 pKa = 10.06GIVMQSSPLFLSAMSRR399 pKa = 11.84LFYY402 pKa = 10.61VLKK405 pKa = 10.64SKK407 pKa = 10.19IHH409 pKa = 5.86IPSGKK414 pKa = 7.2WHH416 pKa = 6.3QLFTLDD422 pKa = 3.84ATAFDD427 pKa = 3.62AAQWFKK433 pKa = 11.14EE434 pKa = 3.85VDD436 pKa = 3.72FSKK439 pKa = 10.64FDD441 pKa = 3.3KK442 pKa = 11.19SQGEE446 pKa = 4.19LHH448 pKa = 6.92HH449 pKa = 6.79KK450 pKa = 7.34VQKK453 pKa = 10.9LIFDD457 pKa = 4.63LLKK460 pKa = 10.74LPPEE464 pKa = 4.1FVEE467 pKa = 4.27MWFTSHH473 pKa = 5.89EE474 pKa = 4.18RR475 pKa = 11.84SHH477 pKa = 6.0ITDD480 pKa = 3.49RR481 pKa = 11.84DD482 pKa = 3.68TGVGFSVDD490 pKa = 3.5YY491 pKa = 10.5QRR493 pKa = 11.84RR494 pKa = 11.84TGDD497 pKa = 3.07ANTYY501 pKa = 10.16LGNTLVTLICLARR514 pKa = 11.84VYY516 pKa = 10.32DD517 pKa = 3.84LCDD520 pKa = 3.56PKK522 pKa = 10.23ITFIIASGDD531 pKa = 3.41DD532 pKa = 3.58SLIGSVEE539 pKa = 4.01EE540 pKa = 4.07LPRR543 pKa = 11.84APEE546 pKa = 3.79HH547 pKa = 6.87LFTSLFNFEE556 pKa = 5.0AKK558 pKa = 10.15FPHH561 pKa = 5.93NQPFICSKK569 pKa = 10.53FLVSVDD575 pKa = 3.54LADD578 pKa = 4.2GGRR581 pKa = 11.84EE582 pKa = 4.11VIAVPNPAKK591 pKa = 10.65LLIRR595 pKa = 11.84MGRR598 pKa = 11.84RR599 pKa = 11.84DD600 pKa = 4.04CQFQSLDD607 pKa = 3.57DD608 pKa = 5.61LYY610 pKa = 10.81ISWLDD615 pKa = 3.31VIYY618 pKa = 10.35YY619 pKa = 10.13FRR621 pKa = 11.84DD622 pKa = 3.06SRR624 pKa = 11.84VCEE627 pKa = 4.25KK628 pKa = 10.76VAEE631 pKa = 4.15LCAYY635 pKa = 8.96RR636 pKa = 11.84QTRR639 pKa = 11.84RR640 pKa = 11.84SSMYY644 pKa = 10.48LLSALLSLPSCFANLKK660 pKa = 9.99KK661 pKa = 10.77FKK663 pKa = 10.3LLCYY667 pKa = 10.22NLTEE671 pKa = 4.15EE672 pKa = 4.37EE673 pKa = 4.39CLKK676 pKa = 9.77KK677 pKa = 9.36TKK679 pKa = 9.02LTRR682 pKa = 11.84DD683 pKa = 3.28THH685 pKa = 4.86GHH687 pKa = 4.32VEE689 pKa = 3.86KK690 pKa = 10.24EE691 pKa = 3.78QRR693 pKa = 11.84VNGCRR698 pKa = 11.84NKK700 pKa = 10.98GIVQEE705 pKa = 4.65GTAIPAKK712 pKa = 9.98NGRR715 pKa = 11.84RR716 pKa = 11.84VQEE719 pKa = 3.79TSTRR723 pKa = 11.84PNSGFRR729 pKa = 11.84AYY731 pKa = 9.41PQRR734 pKa = 11.84KK735 pKa = 8.74SRR737 pKa = 11.84GCWIPHH743 pKa = 5.51ISKK746 pKa = 9.88VFQAPDD752 pKa = 3.2QMGGCSCAVTRR763 pKa = 11.84KK764 pKa = 9.24SFSSIRR770 pKa = 11.84SAVDD774 pKa = 2.94VSGFPPAGPPSRR786 pKa = 11.84YY787 pKa = 9.69AEE789 pKa = 4.09SGEE792 pKa = 4.2FGDD795 pKa = 4.24GTRR798 pKa = 11.84TGSSYY803 pKa = 11.09AYY805 pKa = 9.76KK806 pKa = 10.53NSRR809 pKa = 11.84PRR811 pKa = 11.84NRR813 pKa = 11.84LRR815 pKa = 11.84AQGVHH820 pKa = 6.13QSRR823 pKa = 11.84CSNTEE828 pKa = 3.78YY829 pKa = 10.48IPSNNRR835 pKa = 11.84RR836 pKa = 11.84STT838 pKa = 3.23
MM1 pKa = 7.63EE2 pKa = 6.42IITNVLDD9 pKa = 3.87LVSTVSLRR17 pKa = 11.84TSLQVHH23 pKa = 6.7ASFDD27 pKa = 3.89LPLSIEE33 pKa = 3.85QSFMIRR39 pKa = 11.84LWLVRR44 pKa = 11.84DD45 pKa = 3.79VVIPTMKK52 pKa = 10.43VYY54 pKa = 10.24WDD56 pKa = 3.54ADD58 pKa = 3.44ALLGRR63 pKa = 11.84LEE65 pKa = 4.69TVFSTDD71 pKa = 3.18PEE73 pKa = 4.22PVVMRR78 pKa = 11.84NSTFHH83 pKa = 7.44DD84 pKa = 3.55IVPEE88 pKa = 3.93YY89 pKa = 10.58VDD91 pKa = 3.68EE92 pKa = 4.87FGEE95 pKa = 4.53VPEE98 pKa = 4.18EE99 pKa = 4.43AYY101 pKa = 10.72LSPEE105 pKa = 3.8EE106 pKa = 4.26RR107 pKa = 11.84EE108 pKa = 4.42EE109 pKa = 3.66IHH111 pKa = 6.87KK112 pKa = 9.54ATFLKK117 pKa = 10.33HH118 pKa = 5.15EE119 pKa = 4.29HH120 pKa = 6.52WGSEE124 pKa = 4.02SSGSSFLDD132 pKa = 4.26EE133 pKa = 4.74IEE135 pKa = 4.36CLYY138 pKa = 11.3DD139 pKa = 3.37EE140 pKa = 4.47TVKK143 pKa = 11.12DD144 pKa = 4.68DD145 pKa = 3.88VMQDD149 pKa = 2.74VCTDD153 pKa = 3.26MPKK156 pKa = 10.4DD157 pKa = 3.93YY158 pKa = 11.12EE159 pKa = 4.43IMWNDD164 pKa = 3.55GAVDD168 pKa = 3.98AVWSSRR174 pKa = 11.84FEE176 pKa = 4.02VEE178 pKa = 4.62EE179 pKa = 3.94PPKK182 pKa = 10.62PKK184 pKa = 9.93FRR186 pKa = 11.84PQMTDD191 pKa = 2.89KK192 pKa = 11.21VCDD195 pKa = 3.48PVIIQDD201 pKa = 4.7AINDD205 pKa = 3.88IFPFHH210 pKa = 7.22QDD212 pKa = 2.3MDD214 pKa = 3.76DD215 pKa = 3.76TYY217 pKa = 10.48FQTWVEE223 pKa = 4.15TQDD226 pKa = 3.07ISLEE230 pKa = 4.18VSKK233 pKa = 10.96CWLDD237 pKa = 3.26ASNFKK242 pKa = 10.96DD243 pKa = 3.78FTKK246 pKa = 10.69GQQTYY251 pKa = 10.4AVPTYY256 pKa = 10.83QSGATSKK263 pKa = 10.53RR264 pKa = 11.84VNTQRR269 pKa = 11.84EE270 pKa = 3.87TLLAVKK276 pKa = 9.35KK277 pKa = 10.43RR278 pKa = 11.84NMNIPEE284 pKa = 4.13LQATFDD290 pKa = 4.13LDD292 pKa = 4.62AEE294 pKa = 4.44VATCFKK300 pKa = 10.54RR301 pKa = 11.84FCTHH305 pKa = 5.53VVDD308 pKa = 4.49IPRR311 pKa = 11.84LKK313 pKa = 10.43RR314 pKa = 11.84LPPMMGTEE322 pKa = 4.05TEE324 pKa = 4.2FFHH327 pKa = 7.94RR328 pKa = 11.84LFGWYY333 pKa = 9.7NPPLSEE339 pKa = 4.06YY340 pKa = 10.36RR341 pKa = 11.84GPLTLQSLDD350 pKa = 3.65KK351 pKa = 10.5YY352 pKa = 8.34MHH354 pKa = 5.5MVKK357 pKa = 9.98TIVKK361 pKa = 8.75PVEE364 pKa = 4.43DD365 pKa = 3.85NSLKK369 pKa = 10.61YY370 pKa = 10.06EE371 pKa = 4.23RR372 pKa = 11.84PLCATITLSKK382 pKa = 10.3KK383 pKa = 10.06GIVMQSSPLFLSAMSRR399 pKa = 11.84LFYY402 pKa = 10.61VLKK405 pKa = 10.64SKK407 pKa = 10.19IHH409 pKa = 5.86IPSGKK414 pKa = 7.2WHH416 pKa = 6.3QLFTLDD422 pKa = 3.84ATAFDD427 pKa = 3.62AAQWFKK433 pKa = 11.14EE434 pKa = 3.85VDD436 pKa = 3.72FSKK439 pKa = 10.64FDD441 pKa = 3.3KK442 pKa = 11.19SQGEE446 pKa = 4.19LHH448 pKa = 6.92HH449 pKa = 6.79KK450 pKa = 7.34VQKK453 pKa = 10.9LIFDD457 pKa = 4.63LLKK460 pKa = 10.74LPPEE464 pKa = 4.1FVEE467 pKa = 4.27MWFTSHH473 pKa = 5.89EE474 pKa = 4.18RR475 pKa = 11.84SHH477 pKa = 6.0ITDD480 pKa = 3.49RR481 pKa = 11.84DD482 pKa = 3.68TGVGFSVDD490 pKa = 3.5YY491 pKa = 10.5QRR493 pKa = 11.84RR494 pKa = 11.84TGDD497 pKa = 3.07ANTYY501 pKa = 10.16LGNTLVTLICLARR514 pKa = 11.84VYY516 pKa = 10.32DD517 pKa = 3.84LCDD520 pKa = 3.56PKK522 pKa = 10.23ITFIIASGDD531 pKa = 3.41DD532 pKa = 3.58SLIGSVEE539 pKa = 4.01EE540 pKa = 4.07LPRR543 pKa = 11.84APEE546 pKa = 3.79HH547 pKa = 6.87LFTSLFNFEE556 pKa = 5.0AKK558 pKa = 10.15FPHH561 pKa = 5.93NQPFICSKK569 pKa = 10.53FLVSVDD575 pKa = 3.54LADD578 pKa = 4.2GGRR581 pKa = 11.84EE582 pKa = 4.11VIAVPNPAKK591 pKa = 10.65LLIRR595 pKa = 11.84MGRR598 pKa = 11.84RR599 pKa = 11.84DD600 pKa = 4.04CQFQSLDD607 pKa = 3.57DD608 pKa = 5.61LYY610 pKa = 10.81ISWLDD615 pKa = 3.31VIYY618 pKa = 10.35YY619 pKa = 10.13FRR621 pKa = 11.84DD622 pKa = 3.06SRR624 pKa = 11.84VCEE627 pKa = 4.25KK628 pKa = 10.76VAEE631 pKa = 4.15LCAYY635 pKa = 8.96RR636 pKa = 11.84QTRR639 pKa = 11.84RR640 pKa = 11.84SSMYY644 pKa = 10.48LLSALLSLPSCFANLKK660 pKa = 9.99KK661 pKa = 10.77FKK663 pKa = 10.3LLCYY667 pKa = 10.22NLTEE671 pKa = 4.15EE672 pKa = 4.37EE673 pKa = 4.39CLKK676 pKa = 9.77KK677 pKa = 9.36TKK679 pKa = 9.02LTRR682 pKa = 11.84DD683 pKa = 3.28THH685 pKa = 4.86GHH687 pKa = 4.32VEE689 pKa = 3.86KK690 pKa = 10.24EE691 pKa = 3.78QRR693 pKa = 11.84VNGCRR698 pKa = 11.84NKK700 pKa = 10.98GIVQEE705 pKa = 4.65GTAIPAKK712 pKa = 9.98NGRR715 pKa = 11.84RR716 pKa = 11.84VQEE719 pKa = 3.79TSTRR723 pKa = 11.84PNSGFRR729 pKa = 11.84AYY731 pKa = 9.41PQRR734 pKa = 11.84KK735 pKa = 8.74SRR737 pKa = 11.84GCWIPHH743 pKa = 5.51ISKK746 pKa = 9.88VFQAPDD752 pKa = 3.2QMGGCSCAVTRR763 pKa = 11.84KK764 pKa = 9.24SFSSIRR770 pKa = 11.84SAVDD774 pKa = 2.94VSGFPPAGPPSRR786 pKa = 11.84YY787 pKa = 9.69AEE789 pKa = 4.09SGEE792 pKa = 4.2FGDD795 pKa = 4.24GTRR798 pKa = 11.84TGSSYY803 pKa = 11.09AYY805 pKa = 9.76KK806 pKa = 10.53NSRR809 pKa = 11.84PRR811 pKa = 11.84NRR813 pKa = 11.84LRR815 pKa = 11.84AQGVHH820 pKa = 6.13QSRR823 pKa = 11.84CSNTEE828 pKa = 3.78YY829 pKa = 10.48IPSNNRR835 pKa = 11.84RR836 pKa = 11.84STT838 pKa = 3.23
Molecular weight: 95.93 kDa
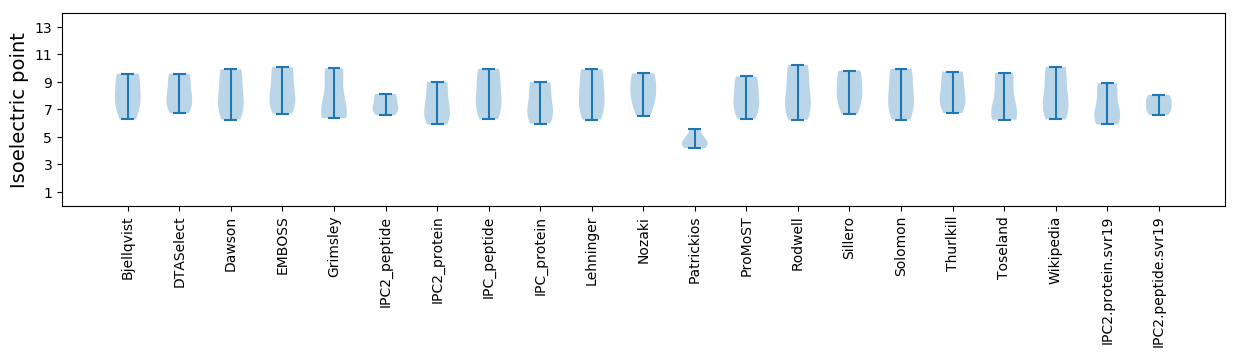
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140GPI7|A0A140GPI7_9BROM Putative silencing suppressor OS=Tomato necrotic streak virus OX=1637492 GN=2b PE=4 SV=1
MM1 pKa = 8.12DD2 pKa = 4.34MSRR5 pKa = 11.84KK6 pKa = 10.22SNVSMDD12 pKa = 3.1AVTRR16 pKa = 11.84EE17 pKa = 4.07LFKK20 pKa = 11.19KK21 pKa = 8.86EE22 pKa = 3.41LRR24 pKa = 11.84YY25 pKa = 9.15RR26 pKa = 11.84LKK28 pKa = 10.13MAEE31 pKa = 4.11EE32 pKa = 4.4FKK34 pKa = 10.98KK35 pKa = 10.29QARR38 pKa = 11.84DD39 pKa = 3.75PIVDD43 pKa = 3.74SVHH46 pKa = 4.81THH48 pKa = 5.19SGNRR52 pKa = 11.84VDD54 pKa = 4.67AGYY57 pKa = 10.14HH58 pKa = 5.44ISPKK62 pKa = 9.97FFKK65 pKa = 10.53PLTKK69 pKa = 9.6WGVVPVLSLVNLSAVSEE86 pKa = 4.23ARR88 pKa = 11.84WMLVDD93 pKa = 3.64SHH95 pKa = 6.64QLAPPLVTPRR105 pKa = 11.84VVSSEE110 pKa = 3.93MAPEE114 pKa = 4.03PEE116 pKa = 3.47ARR118 pKa = 11.84MLIKK122 pKa = 10.37IPDD125 pKa = 3.64LGIDD129 pKa = 3.41YY130 pKa = 9.91EE131 pKa = 4.39LRR133 pKa = 11.84EE134 pKa = 4.25FTNPAVVIQSIYY146 pKa = 10.21RR147 pKa = 11.84RR148 pKa = 11.84IIGEE152 pKa = 4.23VPKK155 pKa = 10.48GWYY158 pKa = 9.02GLKK161 pKa = 9.68SWSISSYY168 pKa = 9.18GDD170 pKa = 3.07VYY172 pKa = 11.38SRR174 pKa = 11.84LRR176 pKa = 11.84VVRR179 pKa = 11.84KK180 pKa = 9.06MKK182 pKa = 10.38VHH184 pKa = 5.65LTIPGSDD191 pKa = 2.94WAYY194 pKa = 9.33TLSLSDD200 pKa = 4.38VISGLAIPRR209 pKa = 11.84LPIPEE214 pKa = 5.36KK215 pKa = 9.55YY216 pKa = 10.55LKK218 pKa = 9.73MPISISFRR226 pKa = 11.84DD227 pKa = 3.68EE228 pKa = 3.63MM229 pKa = 6.3
MM1 pKa = 8.12DD2 pKa = 4.34MSRR5 pKa = 11.84KK6 pKa = 10.22SNVSMDD12 pKa = 3.1AVTRR16 pKa = 11.84EE17 pKa = 4.07LFKK20 pKa = 11.19KK21 pKa = 8.86EE22 pKa = 3.41LRR24 pKa = 11.84YY25 pKa = 9.15RR26 pKa = 11.84LKK28 pKa = 10.13MAEE31 pKa = 4.11EE32 pKa = 4.4FKK34 pKa = 10.98KK35 pKa = 10.29QARR38 pKa = 11.84DD39 pKa = 3.75PIVDD43 pKa = 3.74SVHH46 pKa = 4.81THH48 pKa = 5.19SGNRR52 pKa = 11.84VDD54 pKa = 4.67AGYY57 pKa = 10.14HH58 pKa = 5.44ISPKK62 pKa = 9.97FFKK65 pKa = 10.53PLTKK69 pKa = 9.6WGVVPVLSLVNLSAVSEE86 pKa = 4.23ARR88 pKa = 11.84WMLVDD93 pKa = 3.64SHH95 pKa = 6.64QLAPPLVTPRR105 pKa = 11.84VVSSEE110 pKa = 3.93MAPEE114 pKa = 4.03PEE116 pKa = 3.47ARR118 pKa = 11.84MLIKK122 pKa = 10.37IPDD125 pKa = 3.64LGIDD129 pKa = 3.41YY130 pKa = 9.91EE131 pKa = 4.39LRR133 pKa = 11.84EE134 pKa = 4.25FTNPAVVIQSIYY146 pKa = 10.21RR147 pKa = 11.84RR148 pKa = 11.84IIGEE152 pKa = 4.23VPKK155 pKa = 10.48GWYY158 pKa = 9.02GLKK161 pKa = 9.68SWSISSYY168 pKa = 9.18GDD170 pKa = 3.07VYY172 pKa = 11.38SRR174 pKa = 11.84LRR176 pKa = 11.84VVRR179 pKa = 11.84KK180 pKa = 9.06MKK182 pKa = 10.38VHH184 pKa = 5.65LTIPGSDD191 pKa = 2.94WAYY194 pKa = 9.33TLSLSDD200 pKa = 4.38VISGLAIPRR209 pKa = 11.84LPIPEE214 pKa = 5.36KK215 pKa = 9.55YY216 pKa = 10.55LKK218 pKa = 9.73MPISISFRR226 pKa = 11.84DD227 pKa = 3.68EE228 pKa = 3.63MM229 pKa = 6.3
Molecular weight: 26.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2617 |
213 |
1046 |
523.4 |
59.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.687 ± 0.684 | 2.025 ± 0.298 |
6.19 ± 0.297 | 5.885 ± 0.287 |
4.241 ± 0.601 | 4.7 ± 0.327 |
2.293 ± 0.183 | 5.464 ± 0.434 |
6.458 ± 0.227 | 8.445 ± 0.662 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.981 ± 0.273 | 3.63 ± 0.419 |
5.464 ± 0.484 | 2.828 ± 0.376 |
6.037 ± 0.305 | 8.063 ± 0.362 |
6.305 ± 0.43 | 7.413 ± 0.454 |
1.681 ± 0.15 | 3.21 ± 0.182 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
