
Halomonas sp. SUBG004
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas; unclassified Halomonas
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
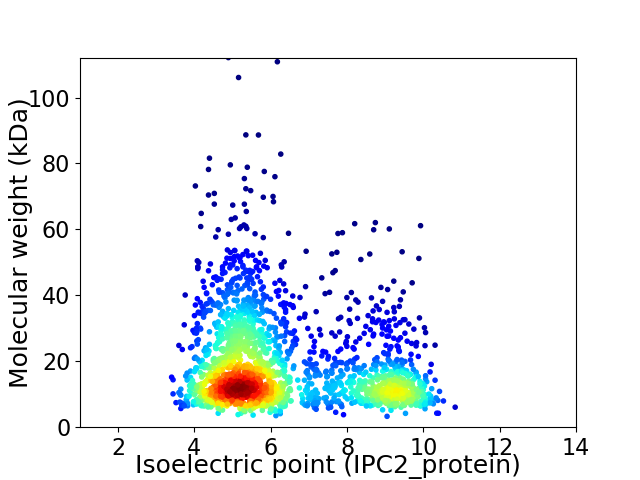
Virtual 2D-PAGE plot for 2023 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
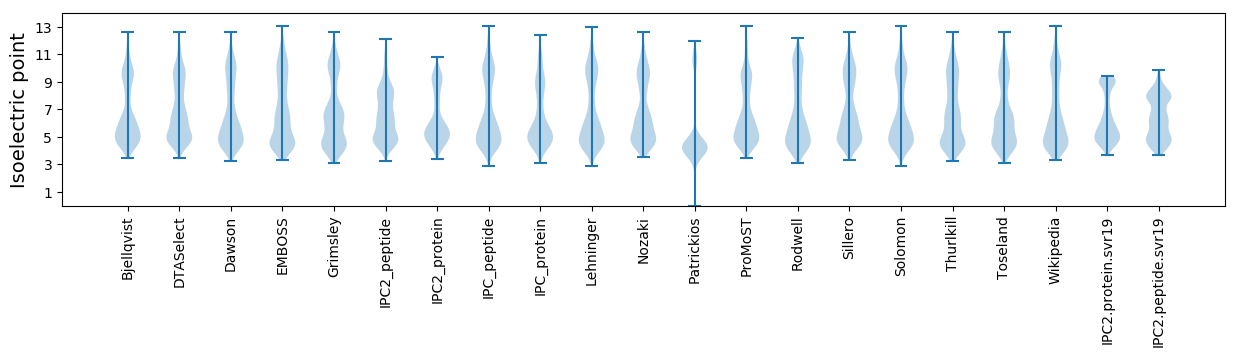
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A085DX94|A0A085DX94_9GAMM Threonylcarbamoyl-AMP synthase OS=Halomonas sp. SUBG004 OX=1485007 GN=tsaC PE=3 SV=1
MM1 pKa = 6.89VWQVEE6 pKa = 4.11DD7 pKa = 4.63SIGNLLNGISNVNAGDD23 pKa = 3.6TAFGDD28 pKa = 3.94DD29 pKa = 4.97AIVNAQTVRR38 pKa = 11.84VTGTTITQAQYY49 pKa = 11.17EE50 pKa = 4.44EE51 pKa = 4.54LLAAVGDD58 pKa = 4.17ALSLEE63 pKa = 4.43AGEE66 pKa = 4.18NAARR70 pKa = 11.84TTVTYY75 pKa = 10.96ADD77 pKa = 3.77LAAAVSAQANGVLAPSDD94 pKa = 4.27FYY96 pKa = 11.62SITANTTYY104 pKa = 10.84TNTNGALSVANALTEE119 pKa = 3.96IEE121 pKa = 4.44TVKK124 pKa = 10.9SILAGARR131 pKa = 11.84NTLNFEE137 pKa = 4.5TVYY140 pKa = 9.97EE141 pKa = 4.0WSITDD146 pKa = 3.53SASNILDD153 pKa = 4.89AITTAVPEE161 pKa = 4.15VLEE164 pKa = 4.32DD165 pKa = 3.6AEE167 pKa = 4.54NIRR170 pKa = 11.84VASGGAGDD178 pKa = 4.81NILAAEE184 pKa = 4.14YY185 pKa = 10.04DD186 pKa = 3.78ALVDD190 pKa = 3.87GLGTIPFDD198 pKa = 4.27YY199 pKa = 11.41NEE201 pKa = 4.01VAYY204 pKa = 9.23TLAYY208 pKa = 10.7VFDD211 pKa = 3.98EE212 pKa = 4.56TNDD215 pKa = 3.31TFAAPDD221 pKa = 3.73DD222 pKa = 3.92LADD225 pKa = 3.82RR226 pKa = 11.84FFFTSDD232 pKa = 2.86VFEE235 pKa = 5.66INDD238 pKa = 3.72ALGWGSMLPRR248 pKa = 11.84QKK250 pKa = 10.66SITMPSRR257 pKa = 11.84RR258 pKa = 11.84WWMQAPTICSSPISTRR274 pKa = 11.84GASWIVRR281 pKa = 11.84PILSMHH287 pKa = 5.68TTATAIGTPPYY298 pKa = 10.73VSDD301 pKa = 3.52ATLVEE306 pKa = 4.39INDD309 pKa = 3.84TAVITFEE316 pKa = 3.94QYY318 pKa = 11.1GILEE322 pKa = 4.28QIPGFEE328 pKa = 4.0IQSYY332 pKa = 6.8TLEE335 pKa = 4.51DD336 pKa = 3.08AVGIYY341 pKa = 10.83LEE343 pKa = 5.24DD344 pKa = 3.68NSGLATNYY352 pKa = 10.09IIDD355 pKa = 3.89TEE357 pKa = 4.25QPYY360 pKa = 9.58QAQKK364 pKa = 11.02SLGRR368 pKa = 11.84RR369 pKa = 11.84CFDD372 pKa = 3.51LL373 pKa = 5.86
MM1 pKa = 6.89VWQVEE6 pKa = 4.11DD7 pKa = 4.63SIGNLLNGISNVNAGDD23 pKa = 3.6TAFGDD28 pKa = 3.94DD29 pKa = 4.97AIVNAQTVRR38 pKa = 11.84VTGTTITQAQYY49 pKa = 11.17EE50 pKa = 4.44EE51 pKa = 4.54LLAAVGDD58 pKa = 4.17ALSLEE63 pKa = 4.43AGEE66 pKa = 4.18NAARR70 pKa = 11.84TTVTYY75 pKa = 10.96ADD77 pKa = 3.77LAAAVSAQANGVLAPSDD94 pKa = 4.27FYY96 pKa = 11.62SITANTTYY104 pKa = 10.84TNTNGALSVANALTEE119 pKa = 3.96IEE121 pKa = 4.44TVKK124 pKa = 10.9SILAGARR131 pKa = 11.84NTLNFEE137 pKa = 4.5TVYY140 pKa = 9.97EE141 pKa = 4.0WSITDD146 pKa = 3.53SASNILDD153 pKa = 4.89AITTAVPEE161 pKa = 4.15VLEE164 pKa = 4.32DD165 pKa = 3.6AEE167 pKa = 4.54NIRR170 pKa = 11.84VASGGAGDD178 pKa = 4.81NILAAEE184 pKa = 4.14YY185 pKa = 10.04DD186 pKa = 3.78ALVDD190 pKa = 3.87GLGTIPFDD198 pKa = 4.27YY199 pKa = 11.41NEE201 pKa = 4.01VAYY204 pKa = 9.23TLAYY208 pKa = 10.7VFDD211 pKa = 3.98EE212 pKa = 4.56TNDD215 pKa = 3.31TFAAPDD221 pKa = 3.73DD222 pKa = 3.92LADD225 pKa = 3.82RR226 pKa = 11.84FFFTSDD232 pKa = 2.86VFEE235 pKa = 5.66INDD238 pKa = 3.72ALGWGSMLPRR248 pKa = 11.84QKK250 pKa = 10.66SITMPSRR257 pKa = 11.84RR258 pKa = 11.84WWMQAPTICSSPISTRR274 pKa = 11.84GASWIVRR281 pKa = 11.84PILSMHH287 pKa = 5.68TTATAIGTPPYY298 pKa = 10.73VSDD301 pKa = 3.52ATLVEE306 pKa = 4.39INDD309 pKa = 3.84TAVITFEE316 pKa = 3.94QYY318 pKa = 11.1GILEE322 pKa = 4.28QIPGFEE328 pKa = 4.0IQSYY332 pKa = 6.8TLEE335 pKa = 4.51DD336 pKa = 3.08AVGIYY341 pKa = 10.83LEE343 pKa = 5.24DD344 pKa = 3.68NSGLATNYY352 pKa = 10.09IIDD355 pKa = 3.89TEE357 pKa = 4.25QPYY360 pKa = 9.58QAQKK364 pKa = 11.02SLGRR368 pKa = 11.84RR369 pKa = 11.84CFDD372 pKa = 3.51LL373 pKa = 5.86
Molecular weight: 40.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085DXN7|A0A085DXN7_9GAMM Glutamate racemase OS=Halomonas sp. SUBG004 OX=1485007 GN=murI PE=3 SV=1
MM1 pKa = 7.2SVVGSTLIGGAIGAWRR17 pKa = 11.84ATAVTGPLPLAVTRR31 pKa = 11.84FPLAKK36 pKa = 10.17LPAAMMGGRR45 pKa = 11.84GRR47 pKa = 11.84WPNRR51 pKa = 11.84LAHH54 pKa = 5.9GHH56 pKa = 6.82LIVV59 pKa = 4.32
MM1 pKa = 7.2SVVGSTLIGGAIGAWRR17 pKa = 11.84ATAVTGPLPLAVTRR31 pKa = 11.84FPLAKK36 pKa = 10.17LPAAMMGGRR45 pKa = 11.84GRR47 pKa = 11.84WPNRR51 pKa = 11.84LAHH54 pKa = 5.9GHH56 pKa = 6.82LIVV59 pKa = 4.32
Molecular weight: 6.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
363457 |
30 |
1032 |
179.7 |
19.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.968 ± 0.063 | 1.058 ± 0.02 |
5.339 ± 0.052 | 6.106 ± 0.063 |
3.492 ± 0.04 | 7.815 ± 0.06 |
2.538 ± 0.035 | 4.675 ± 0.045 |
2.885 ± 0.05 | 11.207 ± 0.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.787 ± 0.031 | 2.813 ± 0.037 |
4.858 ± 0.045 | 4.54 ± 0.057 |
6.519 ± 0.061 | 5.912 ± 0.045 |
5.353 ± 0.043 | 7.177 ± 0.055 |
1.551 ± 0.03 | 2.395 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
