
Azospirillum thermophilum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Azospirillaceae; Azospirillum
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
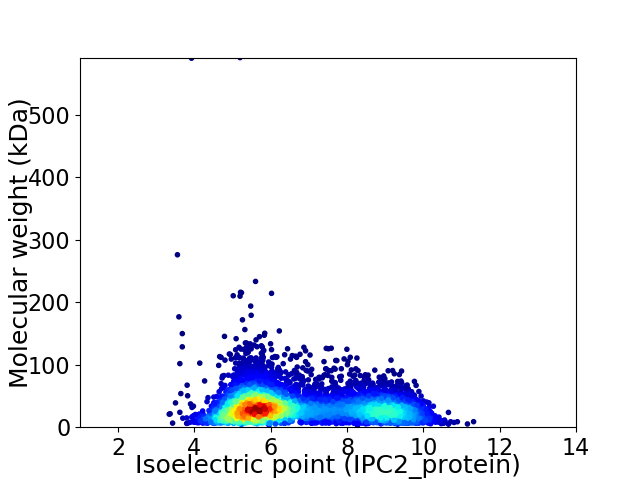
Virtual 2D-PAGE plot for 5547 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
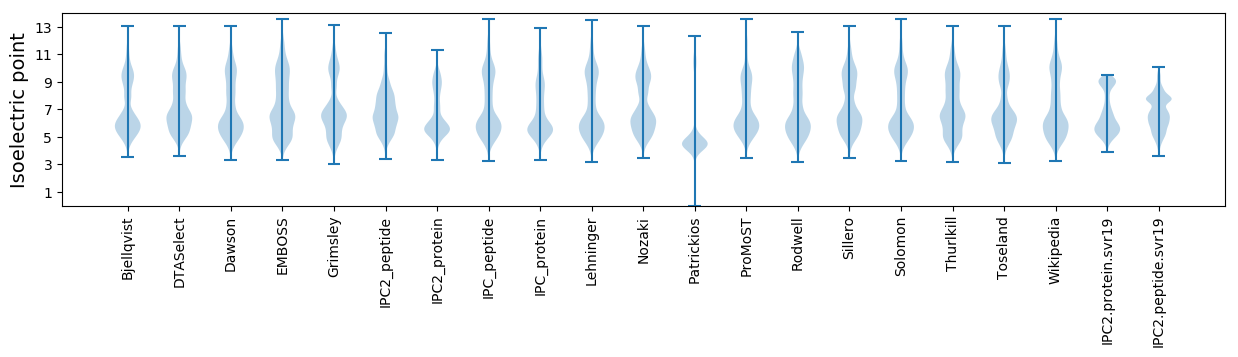
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S2CY96|A0A2S2CY96_9PROT Aryl-sulfate sulfotransferase OS=Azospirillum thermophilum OX=2202148 GN=DEW08_25700 PE=4 SV=1
MM1 pKa = 7.68LGTPGNDD8 pKa = 3.1TLYY11 pKa = 9.52GTSGNDD17 pKa = 3.35LLEE20 pKa = 4.48GLTGNDD26 pKa = 3.3ALYY29 pKa = 10.94GFAGNDD35 pKa = 3.5TLIGGDD41 pKa = 4.12GDD43 pKa = 5.48DD44 pKa = 4.38ILDD47 pKa = 4.36GGSGDD52 pKa = 3.93DD53 pKa = 4.3SMVGGAGNDD62 pKa = 3.51VYY64 pKa = 10.97IVDD67 pKa = 4.36SNSDD71 pKa = 4.03RR72 pKa = 11.84ITEE75 pKa = 4.06LAGEE79 pKa = 4.92GIDD82 pKa = 3.87EE83 pKa = 4.38VRR85 pKa = 11.84TSLSSYY91 pKa = 10.89SLASTPDD98 pKa = 3.56VEE100 pKa = 4.35NLTYY104 pKa = 10.14TGPGAFRR111 pKa = 11.84GTGNALANHH120 pKa = 6.86IKK122 pKa = 10.71GKK124 pKa = 10.34DD125 pKa = 3.62GNDD128 pKa = 3.38TLEE131 pKa = 4.29GAKK134 pKa = 10.84GEE136 pKa = 4.19DD137 pKa = 3.6TLEE140 pKa = 4.37GGNGNDD146 pKa = 2.7IYY148 pKa = 10.45MVYY151 pKa = 10.8GEE153 pKa = 4.71GVTIVEE159 pKa = 4.18AAGAGIDD166 pKa = 3.74EE167 pKa = 4.96VYY169 pKa = 7.77TTLSTYY175 pKa = 7.38TLPDD179 pKa = 3.33GVEE182 pKa = 3.85KK183 pKa = 10.02LTYY186 pKa = 9.11TGNGPFTGIGNALNNRR202 pKa = 11.84LWGPEE207 pKa = 3.5GSTLAGGGGDD217 pKa = 4.06DD218 pKa = 5.62LYY220 pKa = 10.79IISDD224 pKa = 3.46KK225 pKa = 10.75NIRR228 pKa = 11.84IIEE231 pKa = 4.13KK232 pKa = 9.65AGEE235 pKa = 4.6GIDD238 pKa = 4.25EE239 pKa = 4.44IRR241 pKa = 11.84TSLTEE246 pKa = 3.54YY247 pKa = 10.18TLADD251 pKa = 3.55KK252 pKa = 11.42EE253 pKa = 4.3EE254 pKa = 4.3VEE256 pKa = 4.18NLTYY260 pKa = 10.85AGTDD264 pKa = 2.45IVTFIGTGNLKK275 pKa = 10.86NNVITSGSGNDD286 pKa = 3.59TLDD289 pKa = 3.48GGGGIDD295 pKa = 3.93TLSGGAGDD303 pKa = 3.98DD304 pKa = 3.53VYY306 pKa = 11.7YY307 pKa = 10.84VDD309 pKa = 5.82DD310 pKa = 3.9VADD313 pKa = 3.65VVFEE317 pKa = 4.02QVNSGFDD324 pKa = 3.28EE325 pKa = 4.14VRR327 pKa = 11.84ARR329 pKa = 11.84VANYY333 pKa = 10.12SLSNNLEE340 pKa = 3.66QLTFIGSGAFNGTGNSLANVITGGDD365 pKa = 3.82GNDD368 pKa = 3.52TLTGGSGNDD377 pKa = 3.46TLLGAGGSDD386 pKa = 3.36NLIGGDD392 pKa = 3.95GNDD395 pKa = 3.46VLLGGDD401 pKa = 4.3GNDD404 pKa = 3.72MLDD407 pKa = 3.59GGTGNDD413 pKa = 3.89SLDD416 pKa = 3.62GGTGGDD422 pKa = 4.07SFDD425 pKa = 4.46GGAGDD430 pKa = 5.74DD431 pKa = 4.0IYY433 pKa = 11.89VVDD436 pKa = 4.24NVNDD440 pKa = 3.4IVNEE444 pKa = 4.06VGGGGNDD451 pKa = 3.53TVRR454 pKa = 11.84TSLDD458 pKa = 3.66SYY460 pKa = 11.69KK461 pKa = 9.84LTPWVEE467 pKa = 3.87TLIYY471 pKa = 9.84TGGSAFTGTGNDD483 pKa = 3.19LANRR487 pKa = 11.84LEE489 pKa = 4.29GRR491 pKa = 11.84DD492 pKa = 4.0GNDD495 pKa = 3.15TLDD498 pKa = 3.56GRR500 pKa = 11.84EE501 pKa = 4.17GSDD504 pKa = 3.3TLVGGAGDD512 pKa = 3.8DD513 pKa = 3.92VYY515 pKa = 11.06IVDD518 pKa = 4.7SSADD522 pKa = 3.8TIIEE526 pKa = 4.13MANGGNDD533 pKa = 3.44TVRR536 pKa = 11.84TSLSSYY542 pKa = 10.05TLGEE546 pKa = 4.06TSNLEE551 pKa = 3.66NLAFIGSGAFTGTGNSLANAITGGVLSDD579 pKa = 3.91TLSGGAGNDD588 pKa = 3.7TLDD591 pKa = 3.93GGAGNDD597 pKa = 3.88SLIGGAGDD605 pKa = 3.65DD606 pKa = 4.02VYY608 pKa = 11.06IVDD611 pKa = 4.16SSADD615 pKa = 3.24IVVEE619 pKa = 4.01IANGGIDD626 pKa = 3.84TVCTSLGSYY635 pKa = 8.8TLGEE639 pKa = 4.18TSNLEE644 pKa = 3.66NLAFIGSGAFTGTGNSLANAIAGGSGNDD672 pKa = 3.79SLSGGAGNDD681 pKa = 3.46TLTGGLGNDD690 pKa = 4.03TLDD693 pKa = 3.86GGAGNDD699 pKa = 3.88SLIGGAGDD707 pKa = 3.65DD708 pKa = 4.02VYY710 pKa = 11.06IVDD713 pKa = 4.16SSADD717 pKa = 3.3IVVEE721 pKa = 4.06TANGGIDD728 pKa = 3.49TVRR731 pKa = 11.84TSLGSYY737 pKa = 8.91TLGEE741 pKa = 4.18TSNLEE746 pKa = 3.66NLAFIGSGAFTGTGNSLANAIAGGSGNDD774 pKa = 3.69NLSGGAGNDD783 pKa = 3.7TLDD786 pKa = 3.93GGAGNDD792 pKa = 3.7SLIGGVGDD800 pKa = 4.04DD801 pKa = 3.99VYY803 pKa = 11.07IVDD806 pKa = 4.16SSADD810 pKa = 3.24IVVEE814 pKa = 4.01IANGGIDD821 pKa = 3.57TVRR824 pKa = 11.84TSLSGYY830 pKa = 7.68TLGEE834 pKa = 3.97TSNLEE839 pKa = 3.58NLAYY843 pKa = 10.13FGSGAFTGTGNSLANAIAGGSGNDD867 pKa = 3.67SLSGGDD873 pKa = 4.41GNDD876 pKa = 3.34TLTGGLGNDD885 pKa = 3.86TLIGGAGEE893 pKa = 4.12DD894 pKa = 3.59TAVFAAGTIVWRR906 pKa = 11.84DD907 pKa = 2.66IDD909 pKa = 3.62GTEE912 pKa = 4.06RR913 pKa = 11.84AFGPDD918 pKa = 3.47GEE920 pKa = 5.09DD921 pKa = 3.04VLSGIEE927 pKa = 3.93RR928 pKa = 11.84VRR930 pKa = 11.84IGAGAAVPIEE940 pKa = 4.05QSRR943 pKa = 11.84FNPWAYY949 pKa = 10.13LASYY953 pKa = 10.56SDD955 pKa = 3.5LRR957 pKa = 11.84AAFGSDD963 pKa = 2.52GAAAVRR969 pKa = 11.84HH970 pKa = 4.65YY971 pKa = 11.09LEE973 pKa = 4.19YY974 pKa = 11.14GRR976 pKa = 11.84DD977 pKa = 3.38EE978 pKa = 4.08GRR980 pKa = 11.84GIIFDD985 pKa = 4.05TLSYY989 pKa = 8.46TASYY993 pKa = 10.52SDD995 pKa = 4.89LIGAFGTNVDD1005 pKa = 3.35AAARR1009 pKa = 11.84HH1010 pKa = 5.03YY1011 pKa = 10.61LEE1013 pKa = 5.13HH1014 pKa = 6.53GRR1016 pKa = 11.84SEE1018 pKa = 3.92GRR1020 pKa = 11.84GIVFDD1025 pKa = 3.13AHH1027 pKa = 7.06RR1028 pKa = 11.84YY1029 pKa = 7.16LDD1031 pKa = 3.72KK1032 pKa = 11.53YY1033 pKa = 11.32ADD1035 pKa = 3.58LKK1037 pKa = 11.37AAFGDD1042 pKa = 3.81DD1043 pKa = 3.09TVAATRR1049 pKa = 11.84QFILEE1054 pKa = 4.75GYY1056 pKa = 8.42AQGRR1060 pKa = 11.84NLEE1063 pKa = 4.1AAVVNGTAAAEE1074 pKa = 4.08ILQGTGIEE1082 pKa = 4.49DD1083 pKa = 4.59IITGGLGNDD1092 pKa = 3.81TLIGGAGEE1100 pKa = 4.12DD1101 pKa = 3.59TAVFAAGTIVWRR1113 pKa = 11.84DD1114 pKa = 2.66IDD1116 pKa = 3.62GTEE1119 pKa = 4.06RR1120 pKa = 11.84AFGPDD1125 pKa = 3.47GEE1127 pKa = 5.09DD1128 pKa = 3.04VLSGIEE1134 pKa = 3.93RR1135 pKa = 11.84VRR1137 pKa = 11.84IGAGPAVPIEE1147 pKa = 4.07QSRR1150 pKa = 11.84FNPWAYY1156 pKa = 10.13LASYY1160 pKa = 10.56SDD1162 pKa = 3.5LRR1164 pKa = 11.84AAFGSDD1170 pKa = 2.75GAAAVHH1176 pKa = 6.79HH1177 pKa = 5.43YY1178 pKa = 10.79LEE1180 pKa = 4.4YY1181 pKa = 11.17GRR1183 pKa = 11.84DD1184 pKa = 3.38EE1185 pKa = 4.08GRR1187 pKa = 11.84GIIFDD1192 pKa = 4.04ALSYY1196 pKa = 8.27TASYY1200 pKa = 10.62SDD1202 pKa = 4.89LIGAFGTDD1210 pKa = 3.05AEE1212 pKa = 4.62AAAHH1216 pKa = 6.6HH1217 pKa = 5.84YY1218 pKa = 10.42LSYY1221 pKa = 10.98GLRR1224 pKa = 11.84EE1225 pKa = 4.0GRR1227 pKa = 11.84GITFDD1232 pKa = 3.27AQHH1235 pKa = 6.15YY1236 pKa = 8.59LDD1238 pKa = 5.29RR1239 pKa = 11.84YY1240 pKa = 10.26ADD1242 pKa = 3.79LKK1244 pKa = 11.3AAFGNDD1250 pKa = 2.56TAAATRR1256 pKa = 11.84HH1257 pKa = 5.15YY1258 pKa = 9.7IQYY1261 pKa = 10.64GFFEE1265 pKa = 4.53GRR1267 pKa = 11.84LTT1269 pKa = 3.89
MM1 pKa = 7.68LGTPGNDD8 pKa = 3.1TLYY11 pKa = 9.52GTSGNDD17 pKa = 3.35LLEE20 pKa = 4.48GLTGNDD26 pKa = 3.3ALYY29 pKa = 10.94GFAGNDD35 pKa = 3.5TLIGGDD41 pKa = 4.12GDD43 pKa = 5.48DD44 pKa = 4.38ILDD47 pKa = 4.36GGSGDD52 pKa = 3.93DD53 pKa = 4.3SMVGGAGNDD62 pKa = 3.51VYY64 pKa = 10.97IVDD67 pKa = 4.36SNSDD71 pKa = 4.03RR72 pKa = 11.84ITEE75 pKa = 4.06LAGEE79 pKa = 4.92GIDD82 pKa = 3.87EE83 pKa = 4.38VRR85 pKa = 11.84TSLSSYY91 pKa = 10.89SLASTPDD98 pKa = 3.56VEE100 pKa = 4.35NLTYY104 pKa = 10.14TGPGAFRR111 pKa = 11.84GTGNALANHH120 pKa = 6.86IKK122 pKa = 10.71GKK124 pKa = 10.34DD125 pKa = 3.62GNDD128 pKa = 3.38TLEE131 pKa = 4.29GAKK134 pKa = 10.84GEE136 pKa = 4.19DD137 pKa = 3.6TLEE140 pKa = 4.37GGNGNDD146 pKa = 2.7IYY148 pKa = 10.45MVYY151 pKa = 10.8GEE153 pKa = 4.71GVTIVEE159 pKa = 4.18AAGAGIDD166 pKa = 3.74EE167 pKa = 4.96VYY169 pKa = 7.77TTLSTYY175 pKa = 7.38TLPDD179 pKa = 3.33GVEE182 pKa = 3.85KK183 pKa = 10.02LTYY186 pKa = 9.11TGNGPFTGIGNALNNRR202 pKa = 11.84LWGPEE207 pKa = 3.5GSTLAGGGGDD217 pKa = 4.06DD218 pKa = 5.62LYY220 pKa = 10.79IISDD224 pKa = 3.46KK225 pKa = 10.75NIRR228 pKa = 11.84IIEE231 pKa = 4.13KK232 pKa = 9.65AGEE235 pKa = 4.6GIDD238 pKa = 4.25EE239 pKa = 4.44IRR241 pKa = 11.84TSLTEE246 pKa = 3.54YY247 pKa = 10.18TLADD251 pKa = 3.55KK252 pKa = 11.42EE253 pKa = 4.3EE254 pKa = 4.3VEE256 pKa = 4.18NLTYY260 pKa = 10.85AGTDD264 pKa = 2.45IVTFIGTGNLKK275 pKa = 10.86NNVITSGSGNDD286 pKa = 3.59TLDD289 pKa = 3.48GGGGIDD295 pKa = 3.93TLSGGAGDD303 pKa = 3.98DD304 pKa = 3.53VYY306 pKa = 11.7YY307 pKa = 10.84VDD309 pKa = 5.82DD310 pKa = 3.9VADD313 pKa = 3.65VVFEE317 pKa = 4.02QVNSGFDD324 pKa = 3.28EE325 pKa = 4.14VRR327 pKa = 11.84ARR329 pKa = 11.84VANYY333 pKa = 10.12SLSNNLEE340 pKa = 3.66QLTFIGSGAFNGTGNSLANVITGGDD365 pKa = 3.82GNDD368 pKa = 3.52TLTGGSGNDD377 pKa = 3.46TLLGAGGSDD386 pKa = 3.36NLIGGDD392 pKa = 3.95GNDD395 pKa = 3.46VLLGGDD401 pKa = 4.3GNDD404 pKa = 3.72MLDD407 pKa = 3.59GGTGNDD413 pKa = 3.89SLDD416 pKa = 3.62GGTGGDD422 pKa = 4.07SFDD425 pKa = 4.46GGAGDD430 pKa = 5.74DD431 pKa = 4.0IYY433 pKa = 11.89VVDD436 pKa = 4.24NVNDD440 pKa = 3.4IVNEE444 pKa = 4.06VGGGGNDD451 pKa = 3.53TVRR454 pKa = 11.84TSLDD458 pKa = 3.66SYY460 pKa = 11.69KK461 pKa = 9.84LTPWVEE467 pKa = 3.87TLIYY471 pKa = 9.84TGGSAFTGTGNDD483 pKa = 3.19LANRR487 pKa = 11.84LEE489 pKa = 4.29GRR491 pKa = 11.84DD492 pKa = 4.0GNDD495 pKa = 3.15TLDD498 pKa = 3.56GRR500 pKa = 11.84EE501 pKa = 4.17GSDD504 pKa = 3.3TLVGGAGDD512 pKa = 3.8DD513 pKa = 3.92VYY515 pKa = 11.06IVDD518 pKa = 4.7SSADD522 pKa = 3.8TIIEE526 pKa = 4.13MANGGNDD533 pKa = 3.44TVRR536 pKa = 11.84TSLSSYY542 pKa = 10.05TLGEE546 pKa = 4.06TSNLEE551 pKa = 3.66NLAFIGSGAFTGTGNSLANAITGGVLSDD579 pKa = 3.91TLSGGAGNDD588 pKa = 3.7TLDD591 pKa = 3.93GGAGNDD597 pKa = 3.88SLIGGAGDD605 pKa = 3.65DD606 pKa = 4.02VYY608 pKa = 11.06IVDD611 pKa = 4.16SSADD615 pKa = 3.24IVVEE619 pKa = 4.01IANGGIDD626 pKa = 3.84TVCTSLGSYY635 pKa = 8.8TLGEE639 pKa = 4.18TSNLEE644 pKa = 3.66NLAFIGSGAFTGTGNSLANAIAGGSGNDD672 pKa = 3.79SLSGGAGNDD681 pKa = 3.46TLTGGLGNDD690 pKa = 4.03TLDD693 pKa = 3.86GGAGNDD699 pKa = 3.88SLIGGAGDD707 pKa = 3.65DD708 pKa = 4.02VYY710 pKa = 11.06IVDD713 pKa = 4.16SSADD717 pKa = 3.3IVVEE721 pKa = 4.06TANGGIDD728 pKa = 3.49TVRR731 pKa = 11.84TSLGSYY737 pKa = 8.91TLGEE741 pKa = 4.18TSNLEE746 pKa = 3.66NLAFIGSGAFTGTGNSLANAIAGGSGNDD774 pKa = 3.69NLSGGAGNDD783 pKa = 3.7TLDD786 pKa = 3.93GGAGNDD792 pKa = 3.7SLIGGVGDD800 pKa = 4.04DD801 pKa = 3.99VYY803 pKa = 11.07IVDD806 pKa = 4.16SSADD810 pKa = 3.24IVVEE814 pKa = 4.01IANGGIDD821 pKa = 3.57TVRR824 pKa = 11.84TSLSGYY830 pKa = 7.68TLGEE834 pKa = 3.97TSNLEE839 pKa = 3.58NLAYY843 pKa = 10.13FGSGAFTGTGNSLANAIAGGSGNDD867 pKa = 3.67SLSGGDD873 pKa = 4.41GNDD876 pKa = 3.34TLTGGLGNDD885 pKa = 3.86TLIGGAGEE893 pKa = 4.12DD894 pKa = 3.59TAVFAAGTIVWRR906 pKa = 11.84DD907 pKa = 2.66IDD909 pKa = 3.62GTEE912 pKa = 4.06RR913 pKa = 11.84AFGPDD918 pKa = 3.47GEE920 pKa = 5.09DD921 pKa = 3.04VLSGIEE927 pKa = 3.93RR928 pKa = 11.84VRR930 pKa = 11.84IGAGAAVPIEE940 pKa = 4.05QSRR943 pKa = 11.84FNPWAYY949 pKa = 10.13LASYY953 pKa = 10.56SDD955 pKa = 3.5LRR957 pKa = 11.84AAFGSDD963 pKa = 2.52GAAAVRR969 pKa = 11.84HH970 pKa = 4.65YY971 pKa = 11.09LEE973 pKa = 4.19YY974 pKa = 11.14GRR976 pKa = 11.84DD977 pKa = 3.38EE978 pKa = 4.08GRR980 pKa = 11.84GIIFDD985 pKa = 4.05TLSYY989 pKa = 8.46TASYY993 pKa = 10.52SDD995 pKa = 4.89LIGAFGTNVDD1005 pKa = 3.35AAARR1009 pKa = 11.84HH1010 pKa = 5.03YY1011 pKa = 10.61LEE1013 pKa = 5.13HH1014 pKa = 6.53GRR1016 pKa = 11.84SEE1018 pKa = 3.92GRR1020 pKa = 11.84GIVFDD1025 pKa = 3.13AHH1027 pKa = 7.06RR1028 pKa = 11.84YY1029 pKa = 7.16LDD1031 pKa = 3.72KK1032 pKa = 11.53YY1033 pKa = 11.32ADD1035 pKa = 3.58LKK1037 pKa = 11.37AAFGDD1042 pKa = 3.81DD1043 pKa = 3.09TVAATRR1049 pKa = 11.84QFILEE1054 pKa = 4.75GYY1056 pKa = 8.42AQGRR1060 pKa = 11.84NLEE1063 pKa = 4.1AAVVNGTAAAEE1074 pKa = 4.08ILQGTGIEE1082 pKa = 4.49DD1083 pKa = 4.59IITGGLGNDD1092 pKa = 3.81TLIGGAGEE1100 pKa = 4.12DD1101 pKa = 3.59TAVFAAGTIVWRR1113 pKa = 11.84DD1114 pKa = 2.66IDD1116 pKa = 3.62GTEE1119 pKa = 4.06RR1120 pKa = 11.84AFGPDD1125 pKa = 3.47GEE1127 pKa = 5.09DD1128 pKa = 3.04VLSGIEE1134 pKa = 3.93RR1135 pKa = 11.84VRR1137 pKa = 11.84IGAGPAVPIEE1147 pKa = 4.07QSRR1150 pKa = 11.84FNPWAYY1156 pKa = 10.13LASYY1160 pKa = 10.56SDD1162 pKa = 3.5LRR1164 pKa = 11.84AAFGSDD1170 pKa = 2.75GAAAVHH1176 pKa = 6.79HH1177 pKa = 5.43YY1178 pKa = 10.79LEE1180 pKa = 4.4YY1181 pKa = 11.17GRR1183 pKa = 11.84DD1184 pKa = 3.38EE1185 pKa = 4.08GRR1187 pKa = 11.84GIIFDD1192 pKa = 4.04ALSYY1196 pKa = 8.27TASYY1200 pKa = 10.62SDD1202 pKa = 4.89LIGAFGTDD1210 pKa = 3.05AEE1212 pKa = 4.62AAAHH1216 pKa = 6.6HH1217 pKa = 5.84YY1218 pKa = 10.42LSYY1221 pKa = 10.98GLRR1224 pKa = 11.84EE1225 pKa = 4.0GRR1227 pKa = 11.84GITFDD1232 pKa = 3.27AQHH1235 pKa = 6.15YY1236 pKa = 8.59LDD1238 pKa = 5.29RR1239 pKa = 11.84YY1240 pKa = 10.26ADD1242 pKa = 3.79LKK1244 pKa = 11.3AAFGNDD1250 pKa = 2.56TAAATRR1256 pKa = 11.84HH1257 pKa = 5.15YY1258 pKa = 9.7IQYY1261 pKa = 10.64GFFEE1265 pKa = 4.53GRR1267 pKa = 11.84LTT1269 pKa = 3.89
Molecular weight: 128.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S2CVW3|A0A2S2CVW3_9PROT Uncharacterized protein OS=Azospirillum thermophilum OX=2202148 GN=DEW08_21165 PE=4 SV=1
MM1 pKa = 7.42RR2 pKa = 11.84QPRR5 pKa = 11.84TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PAPARR14 pKa = 11.84PWGKK18 pKa = 9.55RR19 pKa = 11.84SRR21 pKa = 11.84RR22 pKa = 11.84PSPRR26 pKa = 11.84SPAPRR31 pKa = 11.84PSRR34 pKa = 11.84SPCAPRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SLRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84SSRR50 pKa = 11.84PRR52 pKa = 11.84SPPRR56 pKa = 11.84PLSRR60 pKa = 11.84LRR62 pKa = 11.84PRR64 pKa = 11.84HH65 pKa = 5.74SRR67 pKa = 11.84RR68 pKa = 11.84PWQSRR73 pKa = 3.07
MM1 pKa = 7.42RR2 pKa = 11.84QPRR5 pKa = 11.84TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PAPARR14 pKa = 11.84PWGKK18 pKa = 9.55RR19 pKa = 11.84SRR21 pKa = 11.84RR22 pKa = 11.84PSPRR26 pKa = 11.84SPAPRR31 pKa = 11.84PSRR34 pKa = 11.84SPCAPRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SLRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84SSRR50 pKa = 11.84PRR52 pKa = 11.84SPPRR56 pKa = 11.84PLSRR60 pKa = 11.84LRR62 pKa = 11.84PRR64 pKa = 11.84HH65 pKa = 5.74SRR67 pKa = 11.84RR68 pKa = 11.84PWQSRR73 pKa = 3.07
Molecular weight: 8.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1799258 |
26 |
6016 |
324.4 |
34.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.401 ± 0.048 | 0.865 ± 0.012 |
5.66 ± 0.03 | 5.587 ± 0.033 |
3.28 ± 0.021 | 9.107 ± 0.035 |
1.99 ± 0.016 | 4.284 ± 0.024 |
2.57 ± 0.031 | 10.748 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.016 | 2.163 ± 0.021 |
5.684 ± 0.036 | 2.969 ± 0.024 |
8.085 ± 0.035 | 4.851 ± 0.026 |
5.293 ± 0.037 | 7.845 ± 0.029 |
1.291 ± 0.012 | 2.013 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
