
Canary polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Gammapolyomavirus; Serinus canaria polyomavirus 1
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
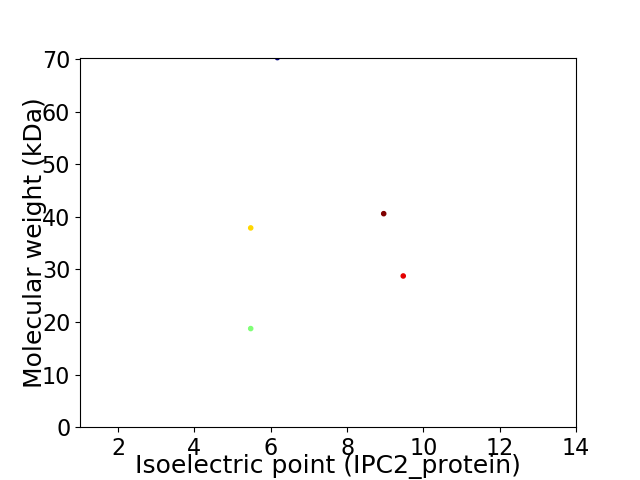
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|E0XL35|E0XL35_9POLY Small t antigen OS=Canary polyomavirus OX=881945 PE=4 SV=1
MM1 pKa = 8.05DD2 pKa = 5.66PGSYY6 pKa = 10.99AEE8 pKa = 5.24LLSLLQLPRR17 pKa = 11.84NADD20 pKa = 3.32EE21 pKa = 4.66KK22 pKa = 11.04DD23 pKa = 2.75ISRR26 pKa = 11.84AYY28 pKa = 10.27RR29 pKa = 11.84KK30 pKa = 9.5VALQMHH36 pKa = 7.18PDD38 pKa = 3.21KK39 pKa = 11.35GGNADD44 pKa = 3.34KK45 pKa = 10.45MKK47 pKa = 10.58RR48 pKa = 11.84LNALMEE54 pKa = 4.25QYY56 pKa = 10.13RR57 pKa = 11.84SSGGSSGPDD66 pKa = 3.23LSCNEE71 pKa = 4.64SPLQSEE77 pKa = 4.99DD78 pKa = 3.68EE79 pKa = 4.49DD80 pKa = 4.48PEE82 pKa = 4.34EE83 pKa = 4.79GPSGEE88 pKa = 4.49SAGSAGYY95 pKa = 9.89FSQSMPRR102 pKa = 11.84SPPVSSQEE110 pKa = 4.01YY111 pKa = 8.21ISAYY115 pKa = 9.39MKK117 pKa = 10.55LLKK120 pKa = 10.49FKK122 pKa = 10.46MSMDD126 pKa = 2.86AFMRR130 pKa = 11.84HH131 pKa = 5.28CEE133 pKa = 3.91RR134 pKa = 11.84RR135 pKa = 11.84KK136 pKa = 9.69QCLTPEE142 pKa = 4.84FGRR145 pKa = 11.84MAEE148 pKa = 4.11NYY150 pKa = 9.56YY151 pKa = 10.42AIPWGVFGRR160 pKa = 11.84LFKK163 pKa = 10.52EE164 pKa = 4.01TDD166 pKa = 2.99LL167 pKa = 5.19
MM1 pKa = 8.05DD2 pKa = 5.66PGSYY6 pKa = 10.99AEE8 pKa = 5.24LLSLLQLPRR17 pKa = 11.84NADD20 pKa = 3.32EE21 pKa = 4.66KK22 pKa = 11.04DD23 pKa = 2.75ISRR26 pKa = 11.84AYY28 pKa = 10.27RR29 pKa = 11.84KK30 pKa = 9.5VALQMHH36 pKa = 7.18PDD38 pKa = 3.21KK39 pKa = 11.35GGNADD44 pKa = 3.34KK45 pKa = 10.45MKK47 pKa = 10.58RR48 pKa = 11.84LNALMEE54 pKa = 4.25QYY56 pKa = 10.13RR57 pKa = 11.84SSGGSSGPDD66 pKa = 3.23LSCNEE71 pKa = 4.64SPLQSEE77 pKa = 4.99DD78 pKa = 3.68EE79 pKa = 4.49DD80 pKa = 4.48PEE82 pKa = 4.34EE83 pKa = 4.79GPSGEE88 pKa = 4.49SAGSAGYY95 pKa = 9.89FSQSMPRR102 pKa = 11.84SPPVSSQEE110 pKa = 4.01YY111 pKa = 8.21ISAYY115 pKa = 9.39MKK117 pKa = 10.55LLKK120 pKa = 10.49FKK122 pKa = 10.46MSMDD126 pKa = 2.86AFMRR130 pKa = 11.84HH131 pKa = 5.28CEE133 pKa = 3.91RR134 pKa = 11.84RR135 pKa = 11.84KK136 pKa = 9.69QCLTPEE142 pKa = 4.84FGRR145 pKa = 11.84MAEE148 pKa = 4.11NYY150 pKa = 9.56YY151 pKa = 10.42AIPWGVFGRR160 pKa = 11.84LFKK163 pKa = 10.52EE164 pKa = 4.01TDD166 pKa = 2.99LL167 pKa = 5.19
Molecular weight: 18.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0XL32|E0XL32_9POLY Minor capsid protein VP2 OS=Canary polyomavirus OX=881945 PE=3 SV=1
MM1 pKa = 7.2GAIVSIFVAIGEE13 pKa = 4.34AVAATGLAAEE23 pKa = 4.86TIISGEE29 pKa = 3.68AAAILGAQISSLAAEE44 pKa = 5.01GFTTAEE50 pKa = 4.13ALSTLGLTPEE60 pKa = 4.75LFAFTTSLASAAEE73 pKa = 4.18SGGALWTGIASGLYY87 pKa = 8.9SLGGAGVSGIAGGLQGGIATAAAGLVRR114 pKa = 11.84FPNISKK120 pKa = 9.54QRR122 pKa = 11.84HH123 pKa = 4.21EE124 pKa = 4.34MALQIWQPLDD134 pKa = 3.0WLYY137 pKa = 10.69PEE139 pKa = 6.0AEE141 pKa = 3.95WLYY144 pKa = 10.94NAYY147 pKa = 10.24HH148 pKa = 6.2YY149 pKa = 10.09FDD151 pKa = 5.15PYY153 pKa = 11.09SWLPSLIDD161 pKa = 3.22QLGYY165 pKa = 10.18YY166 pKa = 9.11FWEE169 pKa = 4.34YY170 pKa = 11.11VEE172 pKa = 4.69RR173 pKa = 11.84TGQRR177 pKa = 11.84QLASQATAVAVRR189 pKa = 11.84ATNRR193 pKa = 11.84AWYY196 pKa = 8.36TLTEE200 pKa = 4.24QISNAGWFIRR210 pKa = 11.84NGYY213 pKa = 8.4YY214 pKa = 10.69ALEE217 pKa = 4.27HH218 pKa = 6.54YY219 pKa = 10.59YY220 pKa = 10.45RR221 pKa = 11.84VLPPRR226 pKa = 11.84FGPPRR231 pKa = 11.84LRR233 pKa = 11.84SYY235 pKa = 11.51LEE237 pKa = 3.8ANRR240 pKa = 11.84EE241 pKa = 4.01NVFIDD246 pKa = 4.09PPDD249 pKa = 3.83MGSEE253 pKa = 3.94TSDD256 pKa = 2.94NRR258 pKa = 11.84FEE260 pKa = 5.2GDD262 pKa = 3.08FLKK265 pKa = 10.64RR266 pKa = 11.84LKK268 pKa = 10.67KK269 pKa = 9.93IFSRR273 pKa = 11.84EE274 pKa = 3.86QEE276 pKa = 4.04SGEE279 pKa = 3.96RR280 pKa = 11.84VYY282 pKa = 11.05KK283 pKa = 10.47SRR285 pKa = 11.84PPGGAEE291 pKa = 4.51QMVTPDD297 pKa = 3.11WLLPLILGLLGDD309 pKa = 4.49LTPTFAADD317 pKa = 2.8IRR319 pKa = 11.84RR320 pKa = 11.84YY321 pKa = 10.08GSEE324 pKa = 3.43NRR326 pKa = 11.84KK327 pKa = 9.58RR328 pKa = 11.84KK329 pKa = 8.4ATSTSTPPVNKK340 pKa = 9.51RR341 pKa = 11.84RR342 pKa = 11.84NRR344 pKa = 11.84GTRR347 pKa = 11.84PKK349 pKa = 10.68DD350 pKa = 3.23RR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84LNNNSRR359 pKa = 11.84GLSKK363 pKa = 10.65SQNGNRR369 pKa = 3.8
MM1 pKa = 7.2GAIVSIFVAIGEE13 pKa = 4.34AVAATGLAAEE23 pKa = 4.86TIISGEE29 pKa = 3.68AAAILGAQISSLAAEE44 pKa = 5.01GFTTAEE50 pKa = 4.13ALSTLGLTPEE60 pKa = 4.75LFAFTTSLASAAEE73 pKa = 4.18SGGALWTGIASGLYY87 pKa = 8.9SLGGAGVSGIAGGLQGGIATAAAGLVRR114 pKa = 11.84FPNISKK120 pKa = 9.54QRR122 pKa = 11.84HH123 pKa = 4.21EE124 pKa = 4.34MALQIWQPLDD134 pKa = 3.0WLYY137 pKa = 10.69PEE139 pKa = 6.0AEE141 pKa = 3.95WLYY144 pKa = 10.94NAYY147 pKa = 10.24HH148 pKa = 6.2YY149 pKa = 10.09FDD151 pKa = 5.15PYY153 pKa = 11.09SWLPSLIDD161 pKa = 3.22QLGYY165 pKa = 10.18YY166 pKa = 9.11FWEE169 pKa = 4.34YY170 pKa = 11.11VEE172 pKa = 4.69RR173 pKa = 11.84TGQRR177 pKa = 11.84QLASQATAVAVRR189 pKa = 11.84ATNRR193 pKa = 11.84AWYY196 pKa = 8.36TLTEE200 pKa = 4.24QISNAGWFIRR210 pKa = 11.84NGYY213 pKa = 8.4YY214 pKa = 10.69ALEE217 pKa = 4.27HH218 pKa = 6.54YY219 pKa = 10.59YY220 pKa = 10.45RR221 pKa = 11.84VLPPRR226 pKa = 11.84FGPPRR231 pKa = 11.84LRR233 pKa = 11.84SYY235 pKa = 11.51LEE237 pKa = 3.8ANRR240 pKa = 11.84EE241 pKa = 4.01NVFIDD246 pKa = 4.09PPDD249 pKa = 3.83MGSEE253 pKa = 3.94TSDD256 pKa = 2.94NRR258 pKa = 11.84FEE260 pKa = 5.2GDD262 pKa = 3.08FLKK265 pKa = 10.64RR266 pKa = 11.84LKK268 pKa = 10.67KK269 pKa = 9.93IFSRR273 pKa = 11.84EE274 pKa = 3.86QEE276 pKa = 4.04SGEE279 pKa = 3.96RR280 pKa = 11.84VYY282 pKa = 11.05KK283 pKa = 10.47SRR285 pKa = 11.84PPGGAEE291 pKa = 4.51QMVTPDD297 pKa = 3.11WLLPLILGLLGDD309 pKa = 4.49LTPTFAADD317 pKa = 2.8IRR319 pKa = 11.84RR320 pKa = 11.84YY321 pKa = 10.08GSEE324 pKa = 3.43NRR326 pKa = 11.84KK327 pKa = 9.58RR328 pKa = 11.84KK329 pKa = 8.4ATSTSTPPVNKK340 pKa = 9.51RR341 pKa = 11.84RR342 pKa = 11.84NRR344 pKa = 11.84GTRR347 pKa = 11.84PKK349 pKa = 10.68DD350 pKa = 3.23RR351 pKa = 11.84RR352 pKa = 11.84RR353 pKa = 11.84LNNNSRR359 pKa = 11.84GLSKK363 pKa = 10.65SQNGNRR369 pKa = 3.8
Molecular weight: 40.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1762 |
167 |
625 |
352.4 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.435 ± 1.2 | 1.589 ± 0.689 |
4.824 ± 0.534 | 6.583 ± 0.546 |
3.292 ± 0.231 | 8.116 ± 1.241 |
1.532 ± 0.351 | 3.462 ± 0.478 |
4.824 ± 0.801 | 10.556 ± 0.546 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.27 ± 0.458 | 5.051 ± 0.368 |
6.47 ± 0.351 | 3.462 ± 0.187 |
6.924 ± 0.949 | 6.924 ± 0.765 |
6.073 ± 0.878 | 5.165 ± 1.091 |
1.249 ± 0.523 | 4.2 ± 0.46 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
