
Cercospora berteroae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Mycosphaerellaceae; Cercospora
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
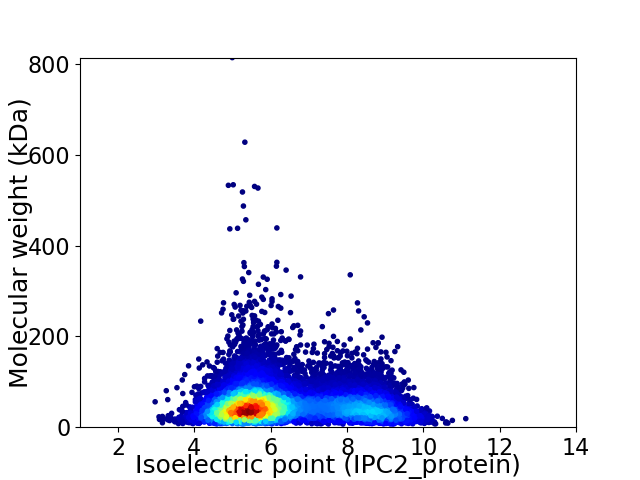
Virtual 2D-PAGE plot for 11903 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2S6BSI7|A0A2S6BSI7_9PEZI Ribokinase OS=Cercospora berteroae OX=357750 GN=CBER1_05560 PE=3 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84SAVRR6 pKa = 11.84GNFWYY11 pKa = 7.13TTSVQQTVATVISTVLDD28 pKa = 3.53YY29 pKa = 11.52GDD31 pKa = 3.85RR32 pKa = 11.84EE33 pKa = 4.25EE34 pKa = 4.46VGNVSTNYY42 pKa = 9.78VPADD46 pKa = 3.26QLYY49 pKa = 8.57TSFGVYY55 pKa = 10.34RR56 pKa = 11.84IRR58 pKa = 11.84TVIFGDD64 pKa = 3.88DD65 pKa = 3.24VVTVNRR71 pKa = 11.84EE72 pKa = 3.88LEE74 pKa = 4.17ALGVPSTLIQGRR86 pKa = 11.84DD87 pKa = 3.32YY88 pKa = 11.46AATIVYY94 pKa = 8.69QANASFDD101 pKa = 3.52HH102 pKa = 6.23GVAYY106 pKa = 7.38PTPFAIWNDD115 pKa = 3.6VQVLYY120 pKa = 11.0LDD122 pKa = 3.83QTTSCIPTATGRR134 pKa = 11.84PTISRR139 pKa = 11.84LEE141 pKa = 4.13GYY143 pKa = 10.34GRR145 pKa = 11.84IDD147 pKa = 3.27IDD149 pKa = 3.19RR150 pKa = 11.84HH151 pKa = 5.02EE152 pKa = 5.11DD153 pKa = 3.09GRR155 pKa = 11.84YY156 pKa = 9.87LSDD159 pKa = 3.38RR160 pKa = 11.84ATKK163 pKa = 10.38YY164 pKa = 10.62YY165 pKa = 7.51YY166 pKa = 10.13TPSAYY171 pKa = 10.15DD172 pKa = 3.27SEE174 pKa = 5.38RR175 pKa = 11.84YY176 pKa = 9.53FYY178 pKa = 11.31VQTLPLDD185 pKa = 4.22GPQPSLGNYY194 pKa = 8.25ALPSGAIEE202 pKa = 4.57RR203 pKa = 11.84YY204 pKa = 10.14VSIQSSAYY212 pKa = 9.42PWITDD217 pKa = 3.61CTPADD222 pKa = 3.94TPGEE226 pKa = 4.25PTVHH230 pKa = 6.43IAVNQLTDD238 pKa = 3.32TSRR241 pKa = 11.84VTVRR245 pKa = 11.84MAGPTTDD252 pKa = 3.4PTPRR256 pKa = 11.84PTGDD260 pKa = 3.15TPALPGTPSPAPPSEE275 pKa = 5.36DD276 pKa = 3.37DD277 pKa = 4.56DD278 pKa = 4.57EE279 pKa = 5.62PIPVQTPTPPTEE291 pKa = 5.4DD292 pKa = 3.59DD293 pKa = 4.39DD294 pKa = 5.9GPVPVQPPANTQVPNSGDD312 pKa = 3.63TPNTGSDD319 pKa = 3.39GSPPQNPTTGEE330 pKa = 3.95QQPANPGEE338 pKa = 4.11TRR340 pKa = 11.84PDD342 pKa = 3.05NSSPNNAGPGGTSFEE357 pKa = 4.27DD358 pKa = 3.74TSPNTQQSPASSNPQSPNPNQNDD381 pKa = 3.37QGPQSDD387 pKa = 4.86DD388 pKa = 3.53SDD390 pKa = 5.69DD391 pKa = 4.16IPSSPAVVVPGEE403 pKa = 4.25LPASGTVQGGAQGQTTAPGSGDD425 pKa = 3.57DD426 pKa = 4.05TSDD429 pKa = 3.22EE430 pKa = 4.42SVPSGTGTDD439 pKa = 3.92DD440 pKa = 3.36QTSGATGSDD449 pKa = 4.04DD450 pKa = 3.75EE451 pKa = 5.75VEE453 pKa = 4.49DD454 pKa = 4.73APASGDD460 pKa = 3.4SSATGDD466 pKa = 3.46VGDD469 pKa = 5.63AIMSGIGQTRR479 pKa = 11.84TGSNGAGYY487 pKa = 8.08TGPAYY492 pKa = 9.5TGKK495 pKa = 10.68APALMANFVVVASMGLTWLAWGCMM519 pKa = 3.77
MM1 pKa = 7.6RR2 pKa = 11.84SAVRR6 pKa = 11.84GNFWYY11 pKa = 7.13TTSVQQTVATVISTVLDD28 pKa = 3.53YY29 pKa = 11.52GDD31 pKa = 3.85RR32 pKa = 11.84EE33 pKa = 4.25EE34 pKa = 4.46VGNVSTNYY42 pKa = 9.78VPADD46 pKa = 3.26QLYY49 pKa = 8.57TSFGVYY55 pKa = 10.34RR56 pKa = 11.84IRR58 pKa = 11.84TVIFGDD64 pKa = 3.88DD65 pKa = 3.24VVTVNRR71 pKa = 11.84EE72 pKa = 3.88LEE74 pKa = 4.17ALGVPSTLIQGRR86 pKa = 11.84DD87 pKa = 3.32YY88 pKa = 11.46AATIVYY94 pKa = 8.69QANASFDD101 pKa = 3.52HH102 pKa = 6.23GVAYY106 pKa = 7.38PTPFAIWNDD115 pKa = 3.6VQVLYY120 pKa = 11.0LDD122 pKa = 3.83QTTSCIPTATGRR134 pKa = 11.84PTISRR139 pKa = 11.84LEE141 pKa = 4.13GYY143 pKa = 10.34GRR145 pKa = 11.84IDD147 pKa = 3.27IDD149 pKa = 3.19RR150 pKa = 11.84HH151 pKa = 5.02EE152 pKa = 5.11DD153 pKa = 3.09GRR155 pKa = 11.84YY156 pKa = 9.87LSDD159 pKa = 3.38RR160 pKa = 11.84ATKK163 pKa = 10.38YY164 pKa = 10.62YY165 pKa = 7.51YY166 pKa = 10.13TPSAYY171 pKa = 10.15DD172 pKa = 3.27SEE174 pKa = 5.38RR175 pKa = 11.84YY176 pKa = 9.53FYY178 pKa = 11.31VQTLPLDD185 pKa = 4.22GPQPSLGNYY194 pKa = 8.25ALPSGAIEE202 pKa = 4.57RR203 pKa = 11.84YY204 pKa = 10.14VSIQSSAYY212 pKa = 9.42PWITDD217 pKa = 3.61CTPADD222 pKa = 3.94TPGEE226 pKa = 4.25PTVHH230 pKa = 6.43IAVNQLTDD238 pKa = 3.32TSRR241 pKa = 11.84VTVRR245 pKa = 11.84MAGPTTDD252 pKa = 3.4PTPRR256 pKa = 11.84PTGDD260 pKa = 3.15TPALPGTPSPAPPSEE275 pKa = 5.36DD276 pKa = 3.37DD277 pKa = 4.56DD278 pKa = 4.57EE279 pKa = 5.62PIPVQTPTPPTEE291 pKa = 5.4DD292 pKa = 3.59DD293 pKa = 4.39DD294 pKa = 5.9GPVPVQPPANTQVPNSGDD312 pKa = 3.63TPNTGSDD319 pKa = 3.39GSPPQNPTTGEE330 pKa = 3.95QQPANPGEE338 pKa = 4.11TRR340 pKa = 11.84PDD342 pKa = 3.05NSSPNNAGPGGTSFEE357 pKa = 4.27DD358 pKa = 3.74TSPNTQQSPASSNPQSPNPNQNDD381 pKa = 3.37QGPQSDD387 pKa = 4.86DD388 pKa = 3.53SDD390 pKa = 5.69DD391 pKa = 4.16IPSSPAVVVPGEE403 pKa = 4.25LPASGTVQGGAQGQTTAPGSGDD425 pKa = 3.57DD426 pKa = 4.05TSDD429 pKa = 3.22EE430 pKa = 4.42SVPSGTGTDD439 pKa = 3.92DD440 pKa = 3.36QTSGATGSDD449 pKa = 4.04DD450 pKa = 3.75EE451 pKa = 5.75VEE453 pKa = 4.49DD454 pKa = 4.73APASGDD460 pKa = 3.4SSATGDD466 pKa = 3.46VGDD469 pKa = 5.63AIMSGIGQTRR479 pKa = 11.84TGSNGAGYY487 pKa = 8.08TGPAYY492 pKa = 9.5TGKK495 pKa = 10.68APALMANFVVVASMGLTWLAWGCMM519 pKa = 3.77
Molecular weight: 54.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S6BSK1|A0A2S6BSK1_9PEZI Uncharacterized protein OS=Cercospora berteroae OX=357750 GN=CBER1_05571 PE=4 SV=1
MM1 pKa = 7.35NGLGPFMSIGRR12 pKa = 11.84GRR14 pKa = 11.84ITMFSPGGRR23 pKa = 11.84GRR25 pKa = 11.84GTQIMTIGGNGFINPAGSSIQIFGNGRR52 pKa = 11.84GGMNIMSNGGRR63 pKa = 11.84GRR65 pKa = 11.84GNYY68 pKa = 10.29SGMQSGTGFNNLGQIQQITMPNPMTGAGGGMFMSMPGMTMSIPFQGGRR116 pKa = 11.84GTGRR120 pKa = 11.84GTGFGLTSAGSSASGSRR137 pKa = 11.84ASGSSRR143 pKa = 11.84SSSSRR148 pKa = 11.84APGSTASGSMGRR160 pKa = 11.84GSSGRR165 pKa = 11.84GSHH168 pKa = 6.58ASSAAGAGSVYY179 pKa = 10.5GGSQGPFGGGGGGTAAPP196 pKa = 4.29
MM1 pKa = 7.35NGLGPFMSIGRR12 pKa = 11.84GRR14 pKa = 11.84ITMFSPGGRR23 pKa = 11.84GRR25 pKa = 11.84GTQIMTIGGNGFINPAGSSIQIFGNGRR52 pKa = 11.84GGMNIMSNGGRR63 pKa = 11.84GRR65 pKa = 11.84GNYY68 pKa = 10.29SGMQSGTGFNNLGQIQQITMPNPMTGAGGGMFMSMPGMTMSIPFQGGRR116 pKa = 11.84GTGRR120 pKa = 11.84GTGFGLTSAGSSASGSRR137 pKa = 11.84ASGSSRR143 pKa = 11.84SSSSRR148 pKa = 11.84APGSTASGSMGRR160 pKa = 11.84GSSGRR165 pKa = 11.84GSHH168 pKa = 6.58ASSAAGAGSVYY179 pKa = 10.5GGSQGPFGGGGGGTAAPP196 pKa = 4.29
Molecular weight: 18.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5823408 |
66 |
7529 |
489.2 |
54.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.195 ± 0.02 | 1.169 ± 0.008 |
5.851 ± 0.015 | 6.358 ± 0.021 |
3.568 ± 0.014 | 6.92 ± 0.022 |
2.422 ± 0.009 | 4.647 ± 0.014 |
4.952 ± 0.021 | 8.585 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.009 | 3.701 ± 0.014 |
5.902 ± 0.025 | 4.327 ± 0.017 |
6.097 ± 0.02 | 7.882 ± 0.025 |
6.047 ± 0.02 | 5.997 ± 0.015 |
1.468 ± 0.009 | 2.753 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
