
Azospirillum lipoferum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Azospirillum
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
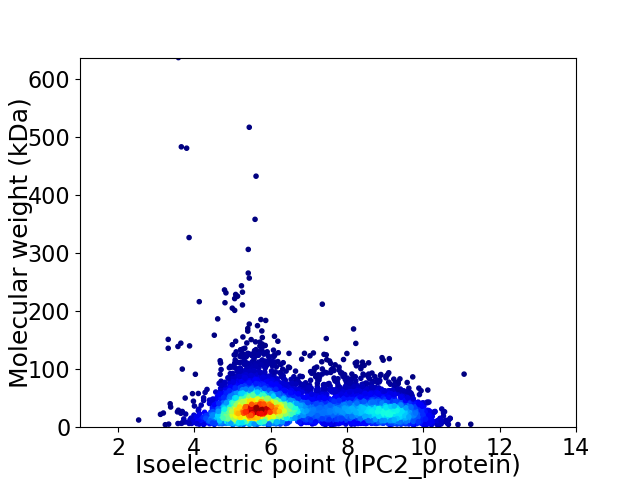
Virtual 2D-PAGE plot for 6618 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7NY64|A0A1X7NY64_AZOLI Plasmid segregation centromere-binding protein ParG OS=Azospirillum lipoferum OX=193 GN=SAMN02982994_1873 PE=4 SV=1
MM1 pKa = 7.04STDD4 pKa = 3.3ATEE7 pKa = 4.51LAPNAAPTPSADD19 pKa = 3.19LAYY22 pKa = 10.59YY23 pKa = 9.86IAALLPVGTPKK34 pKa = 10.16WGVATPGNAASVTFSFMEE52 pKa = 4.35SVPSYY57 pKa = 11.51ADD59 pKa = 3.37AGDD62 pKa = 3.77AAGFAPMNATQRR74 pKa = 11.84AAVRR78 pKa = 11.84QALAAWTEE86 pKa = 4.04VANITFTEE94 pKa = 4.42VADD97 pKa = 4.45GGAGGQLRR105 pKa = 11.84FGTNSQGGVSSGYY118 pKa = 10.59GYY120 pKa = 10.92YY121 pKa = 10.22PNASLAAGGDD131 pKa = 3.6VYY133 pKa = 10.99LAKK136 pKa = 10.4DD137 pKa = 3.43ASSNTSPTAGTYY149 pKa = 9.5GYY151 pKa = 7.94EE152 pKa = 4.37TIVHH156 pKa = 6.9EE157 pKa = 4.54IGHH160 pKa = 6.9AIGLKK165 pKa = 10.61HH166 pKa = 6.53PGNYY170 pKa = 9.14NAGGGGTEE178 pKa = 4.36APYY181 pKa = 10.94LPTAEE186 pKa = 5.88DD187 pKa = 3.51STQYY191 pKa = 11.12TVMSYY196 pKa = 11.19NDD198 pKa = 4.04HH199 pKa = 7.3PSLGLNGLLTGPVLYY214 pKa = 10.49DD215 pKa = 3.52IAAIQYY221 pKa = 10.32LYY223 pKa = 10.85GANTTTRR230 pKa = 11.84SGNTTYY236 pKa = 10.72SYY238 pKa = 11.53NSAATVVSGSIWDD251 pKa = 3.73AGGTDD256 pKa = 3.48TLDD259 pKa = 3.97ASGQTASVVINLAPGSFSSIGPNGSGGRR287 pKa = 11.84AVANLSIAYY296 pKa = 7.38GTTIEE301 pKa = 4.25NANGGSGDD309 pKa = 4.2DD310 pKa = 4.34SISGNAIDD318 pKa = 4.86NVLNGGGGNDD328 pKa = 4.0TISGGGGADD337 pKa = 3.43SIIVNGGIVTVDD349 pKa = 3.17GGSGIDD355 pKa = 3.51TLTIGAIGTVVRR367 pKa = 11.84ATGLEE372 pKa = 3.97NFIGGAGQDD381 pKa = 3.23RR382 pKa = 11.84VILIDD387 pKa = 3.48VGNTIVMSAIDD398 pKa = 3.81TLVGSGGNDD407 pKa = 2.85VVTLGTVANYY417 pKa = 10.57SIIAGIEE424 pKa = 4.02TLIGGAATDD433 pKa = 4.53IIVLANGGGTLIATGIEE450 pKa = 4.17YY451 pKa = 10.67LIGGAGQDD459 pKa = 2.99IAMLAGTGSTLITRR473 pKa = 11.84GIEE476 pKa = 4.04TLVGSTGQDD485 pKa = 2.46IVTLGDD491 pKa = 3.62GGNTMLAFAGLEE503 pKa = 3.93YY504 pKa = 10.59LIGGAGVDD512 pKa = 4.04AVTLGPGGNTIIVRR526 pKa = 11.84GLEE529 pKa = 4.14TLTGGAGNDD538 pKa = 3.03VVTFGNSGNLTTVEE552 pKa = 4.12DD553 pKa = 4.34LEE555 pKa = 4.8TIVGGTGTDD564 pKa = 3.39IILTGARR571 pKa = 11.84GSTLITAGIEE581 pKa = 4.01YY582 pKa = 10.25LIGGAGQDD590 pKa = 2.99IAMLAGTGNTLITRR604 pKa = 11.84GVEE607 pKa = 4.14TLVGSAGQDD616 pKa = 3.31VIMLGDD622 pKa = 4.16GGNTLTAFAAIEE634 pKa = 4.05YY635 pKa = 10.09LVGGAANDD643 pKa = 3.91LVLLGAGGSTVTVRR657 pKa = 11.84GLEE660 pKa = 4.07TLIGGAGIDD669 pKa = 3.88AVTLGNSYY677 pKa = 11.31NFITVGATEE686 pKa = 4.31TLTGGTSTDD695 pKa = 3.02IVVLAAGGSTLIATGLEE712 pKa = 4.12YY713 pKa = 10.8LIGGAGIDD721 pKa = 3.56IVALANGGNTVIVRR735 pKa = 11.84GLEE738 pKa = 3.99TLAGGIGTDD747 pKa = 3.95SLILGNSANFMTIGGIEE764 pKa = 4.36TLTGGTSTDD773 pKa = 2.69IVVLNDD779 pKa = 3.37AGSTMVASGIEE790 pKa = 3.98YY791 pKa = 10.38LIGGGAMDD799 pKa = 4.42VVTLGAAGTLVTRR812 pKa = 11.84GVEE815 pKa = 4.45TIAGSAGMDD824 pKa = 3.44TLILGDD830 pKa = 3.83STNTLTVTGVEE841 pKa = 4.21TLVGGVSADD850 pKa = 3.65TVTVAAGAIRR860 pKa = 11.84FEE862 pKa = 4.61GGGGADD868 pKa = 3.69AVTLAAGQAGDD879 pKa = 3.15RR880 pKa = 11.84VVFRR884 pKa = 11.84SVGDD888 pKa = 3.4GGGPGASVGYY898 pKa = 10.52DD899 pKa = 3.25RR900 pKa = 11.84ITNFGTGDD908 pKa = 3.49AVVVAGQMAGFVDD921 pKa = 5.0RR922 pKa = 11.84NANGALQTAVRR933 pKa = 11.84NTNQINLVTDD943 pKa = 3.7EE944 pKa = 4.71AVLLSVALASLTDD957 pKa = 3.42EE958 pKa = 4.66GFVNLRR964 pKa = 11.84AGIGKK969 pKa = 8.67MAGGVAGISALAVASDD985 pKa = 3.97GVDD988 pKa = 2.81TGVYY992 pKa = 10.37LITDD996 pKa = 3.81GDD998 pKa = 4.07GDD1000 pKa = 3.58GWLAAGEE1007 pKa = 4.12VRR1009 pKa = 11.84LLGLFGAAGIGASQLLAGG1027 pKa = 4.64
MM1 pKa = 7.04STDD4 pKa = 3.3ATEE7 pKa = 4.51LAPNAAPTPSADD19 pKa = 3.19LAYY22 pKa = 10.59YY23 pKa = 9.86IAALLPVGTPKK34 pKa = 10.16WGVATPGNAASVTFSFMEE52 pKa = 4.35SVPSYY57 pKa = 11.51ADD59 pKa = 3.37AGDD62 pKa = 3.77AAGFAPMNATQRR74 pKa = 11.84AAVRR78 pKa = 11.84QALAAWTEE86 pKa = 4.04VANITFTEE94 pKa = 4.42VADD97 pKa = 4.45GGAGGQLRR105 pKa = 11.84FGTNSQGGVSSGYY118 pKa = 10.59GYY120 pKa = 10.92YY121 pKa = 10.22PNASLAAGGDD131 pKa = 3.6VYY133 pKa = 10.99LAKK136 pKa = 10.4DD137 pKa = 3.43ASSNTSPTAGTYY149 pKa = 9.5GYY151 pKa = 7.94EE152 pKa = 4.37TIVHH156 pKa = 6.9EE157 pKa = 4.54IGHH160 pKa = 6.9AIGLKK165 pKa = 10.61HH166 pKa = 6.53PGNYY170 pKa = 9.14NAGGGGTEE178 pKa = 4.36APYY181 pKa = 10.94LPTAEE186 pKa = 5.88DD187 pKa = 3.51STQYY191 pKa = 11.12TVMSYY196 pKa = 11.19NDD198 pKa = 4.04HH199 pKa = 7.3PSLGLNGLLTGPVLYY214 pKa = 10.49DD215 pKa = 3.52IAAIQYY221 pKa = 10.32LYY223 pKa = 10.85GANTTTRR230 pKa = 11.84SGNTTYY236 pKa = 10.72SYY238 pKa = 11.53NSAATVVSGSIWDD251 pKa = 3.73AGGTDD256 pKa = 3.48TLDD259 pKa = 3.97ASGQTASVVINLAPGSFSSIGPNGSGGRR287 pKa = 11.84AVANLSIAYY296 pKa = 7.38GTTIEE301 pKa = 4.25NANGGSGDD309 pKa = 4.2DD310 pKa = 4.34SISGNAIDD318 pKa = 4.86NVLNGGGGNDD328 pKa = 4.0TISGGGGADD337 pKa = 3.43SIIVNGGIVTVDD349 pKa = 3.17GGSGIDD355 pKa = 3.51TLTIGAIGTVVRR367 pKa = 11.84ATGLEE372 pKa = 3.97NFIGGAGQDD381 pKa = 3.23RR382 pKa = 11.84VILIDD387 pKa = 3.48VGNTIVMSAIDD398 pKa = 3.81TLVGSGGNDD407 pKa = 2.85VVTLGTVANYY417 pKa = 10.57SIIAGIEE424 pKa = 4.02TLIGGAATDD433 pKa = 4.53IIVLANGGGTLIATGIEE450 pKa = 4.17YY451 pKa = 10.67LIGGAGQDD459 pKa = 2.99IAMLAGTGSTLITRR473 pKa = 11.84GIEE476 pKa = 4.04TLVGSTGQDD485 pKa = 2.46IVTLGDD491 pKa = 3.62GGNTMLAFAGLEE503 pKa = 3.93YY504 pKa = 10.59LIGGAGVDD512 pKa = 4.04AVTLGPGGNTIIVRR526 pKa = 11.84GLEE529 pKa = 4.14TLTGGAGNDD538 pKa = 3.03VVTFGNSGNLTTVEE552 pKa = 4.12DD553 pKa = 4.34LEE555 pKa = 4.8TIVGGTGTDD564 pKa = 3.39IILTGARR571 pKa = 11.84GSTLITAGIEE581 pKa = 4.01YY582 pKa = 10.25LIGGAGQDD590 pKa = 2.99IAMLAGTGNTLITRR604 pKa = 11.84GVEE607 pKa = 4.14TLVGSAGQDD616 pKa = 3.31VIMLGDD622 pKa = 4.16GGNTLTAFAAIEE634 pKa = 4.05YY635 pKa = 10.09LVGGAANDD643 pKa = 3.91LVLLGAGGSTVTVRR657 pKa = 11.84GLEE660 pKa = 4.07TLIGGAGIDD669 pKa = 3.88AVTLGNSYY677 pKa = 11.31NFITVGATEE686 pKa = 4.31TLTGGTSTDD695 pKa = 3.02IVVLAAGGSTLIATGLEE712 pKa = 4.12YY713 pKa = 10.8LIGGAGIDD721 pKa = 3.56IVALANGGNTVIVRR735 pKa = 11.84GLEE738 pKa = 3.99TLAGGIGTDD747 pKa = 3.95SLILGNSANFMTIGGIEE764 pKa = 4.36TLTGGTSTDD773 pKa = 2.69IVVLNDD779 pKa = 3.37AGSTMVASGIEE790 pKa = 3.98YY791 pKa = 10.38LIGGGAMDD799 pKa = 4.42VVTLGAAGTLVTRR812 pKa = 11.84GVEE815 pKa = 4.45TIAGSAGMDD824 pKa = 3.44TLILGDD830 pKa = 3.83STNTLTVTGVEE841 pKa = 4.21TLVGGVSADD850 pKa = 3.65TVTVAAGAIRR860 pKa = 11.84FEE862 pKa = 4.61GGGGADD868 pKa = 3.69AVTLAAGQAGDD879 pKa = 3.15RR880 pKa = 11.84VVFRR884 pKa = 11.84SVGDD888 pKa = 3.4GGGPGASVGYY898 pKa = 10.52DD899 pKa = 3.25RR900 pKa = 11.84ITNFGTGDD908 pKa = 3.49AVVVAGQMAGFVDD921 pKa = 5.0RR922 pKa = 11.84NANGALQTAVRR933 pKa = 11.84NTNQINLVTDD943 pKa = 3.7EE944 pKa = 4.71AVLLSVALASLTDD957 pKa = 3.42EE958 pKa = 4.66GFVNLRR964 pKa = 11.84AGIGKK969 pKa = 8.67MAGGVAGISALAVASDD985 pKa = 3.97GVDD988 pKa = 2.81TGVYY992 pKa = 10.37LITDD996 pKa = 3.81GDD998 pKa = 4.07GDD1000 pKa = 3.58GWLAAGEE1007 pKa = 4.12VRR1009 pKa = 11.84LLGLFGAAGIGASQLLAGG1027 pKa = 4.64
Molecular weight: 100.06 kDa
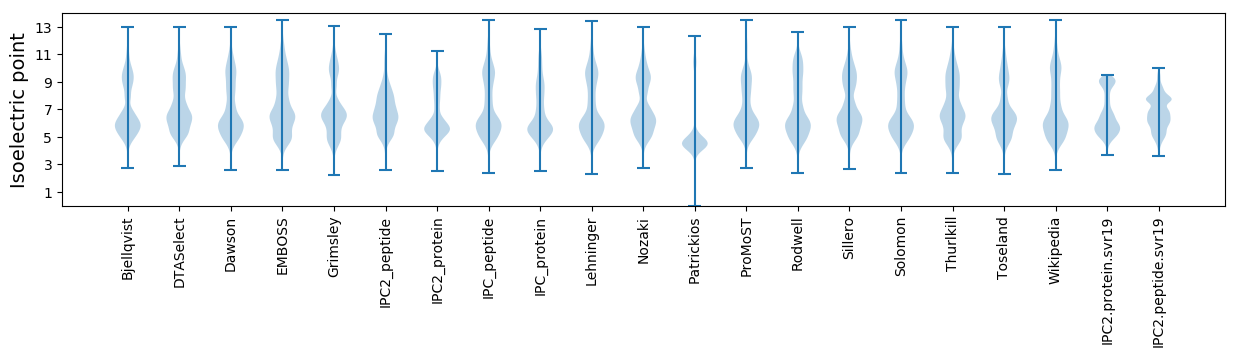
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7Q0R6|A0A1X7Q0R6_AZOLI 2-hydroxychromene-2-carboxylate isomerase OS=Azospirillum lipoferum OX=193 GN=SAMN02982994_4328 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIVRR12 pKa = 11.84KK13 pKa = 9.1HH14 pKa = 4.0RR15 pKa = 11.84HH16 pKa = 4.06GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.3VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIVRR12 pKa = 11.84KK13 pKa = 9.1HH14 pKa = 4.0RR15 pKa = 11.84HH16 pKa = 4.06GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.3VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.91VLSAA44 pKa = 4.11
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2258444 |
39 |
6408 |
341.3 |
36.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.647 ± 0.05 | 0.805 ± 0.01 |
5.787 ± 0.031 | 5.377 ± 0.03 |
3.29 ± 0.019 | 9.247 ± 0.048 |
2.029 ± 0.015 | 4.258 ± 0.026 |
2.567 ± 0.024 | 10.79 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.245 ± 0.017 | 2.281 ± 0.017 |
5.686 ± 0.036 | 2.823 ± 0.019 |
7.881 ± 0.042 | 4.959 ± 0.024 |
5.45 ± 0.043 | 7.673 ± 0.025 |
1.287 ± 0.013 | 1.919 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
