
Deinococcus maricopensis (strain DSM 21211 / LMG 22137 / NRRL B-23946 / LB-34)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus; Deinococcus maricopensis
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
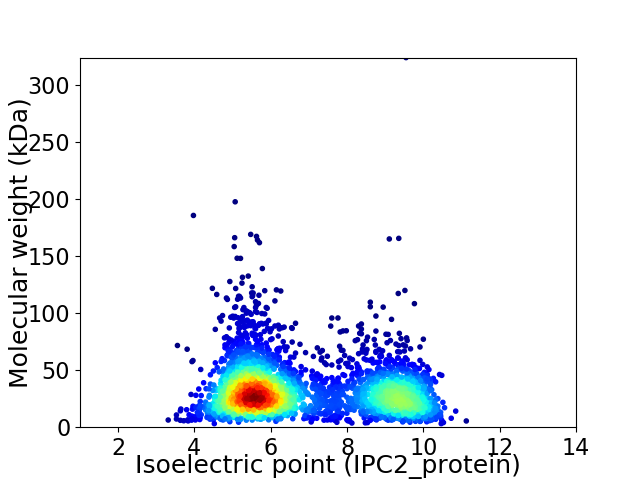
Virtual 2D-PAGE plot for 3262 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E8UC11|E8UC11_DEIML Glycosyl transferase group 1 OS=Deinococcus maricopensis (strain DSM 21211 / LMG 22137 / NRRL B-23946 / LB-34) OX=709986 GN=Deima_2971 PE=4 SV=1
MM1 pKa = 7.41HH2 pKa = 7.73PATRR6 pKa = 11.84TTPALIRR13 pKa = 11.84TLLTLAALLGLTWAPAHH30 pKa = 6.33AVTFNLSFAGGVDD43 pKa = 3.41VTTGSTCATALNSRR57 pKa = 11.84CRR59 pKa = 11.84FNSVVVGAGTGPYY72 pKa = 9.79QRR74 pKa = 11.84DD75 pKa = 3.56AIITISKK82 pKa = 7.98LTNAAINNSSVAAPNSDD99 pKa = 4.16LDD101 pKa = 3.98NPAPTLTGTAPAASAQDD118 pKa = 3.83FFSPTVDD125 pKa = 3.04VSSAGTGTGWAEE137 pKa = 3.53FTIDD141 pKa = 4.17FIARR145 pKa = 11.84GAAVPAVGAGNAALPGTFHH164 pKa = 5.85VTSFDD169 pKa = 3.32TDD171 pKa = 3.07AANPLRR177 pKa = 11.84EE178 pKa = 4.07FVEE181 pKa = 4.75YY182 pKa = 10.39VAPATTALSSGTTLTSRR199 pKa = 11.84AAVDD203 pKa = 3.27GGVAYY208 pKa = 10.53QSGTTGVSGISTDD221 pKa = 4.26PIYY224 pKa = 10.62KK225 pKa = 10.53ASATYY230 pKa = 10.17TNAASVRR237 pKa = 11.84LVYY240 pKa = 10.18GANTGGTACSGATCEE255 pKa = 4.67RR256 pKa = 11.84LTAFDD261 pKa = 5.35FYY263 pKa = 11.55VADD266 pKa = 3.98SVVLQTDD273 pKa = 3.48VNGFKK278 pKa = 10.48SARR281 pKa = 11.84LTTDD285 pKa = 2.79ADD287 pKa = 3.98GNGAITPGDD296 pKa = 3.57TVTYY300 pKa = 9.0TVTYY304 pKa = 10.65VNTGNTASTGFQITDD319 pKa = 3.69ALPSGVTTTAGAQTVRR335 pKa = 11.84LNGTVTAAARR345 pKa = 11.84NTAYY349 pKa = 9.7TGTGANTTLLAAGQTLPANGTISVDD374 pKa = 2.95IPVVVGAVTSDD385 pKa = 3.15NTVLSNQASGSGSGATNVLSDD406 pKa = 3.96NVDD409 pKa = 2.93ATTAFPPSVTGATGFAAPPAGSVPQTQTAAVSPTTITVRR448 pKa = 11.84RR449 pKa = 11.84QPTLTLNKK457 pKa = 9.3TIAAPGRR464 pKa = 11.84VNTNDD469 pKa = 2.96QFTVQISSGGTTVASATTTGTNTTATTGAVTVASGTTYY507 pKa = 10.55TLSEE511 pKa = 4.1ALAAGSGSTLSAYY524 pKa = 10.33ASTMTCTNATGGSSTTLPGGAGVSFTVTPTFGDD557 pKa = 3.86DD558 pKa = 3.59LTCTFTNTGTPVDD571 pKa = 4.15LAVSQTGPATANLDD585 pKa = 3.43GTVTYY590 pKa = 7.97TVRR593 pKa = 11.84VWNNGSDD600 pKa = 3.64VTGATFTDD608 pKa = 4.14TVPAALTNVTWTCAATGSASCGATTSGTGNTISATLGALTTDD650 pKa = 3.42TGSAATADD658 pKa = 3.77TNYY661 pKa = 8.86VTFTITGTATSTGSLTNTATVTAPGGFTEE690 pKa = 5.85SSTTNNSASTSTTVTATVQCSTLYY714 pKa = 9.53GTFGTGRR721 pKa = 11.84EE722 pKa = 4.12LRR724 pKa = 11.84SVTEE728 pKa = 4.12TNTVGSLIGTIPDD741 pKa = 3.21QGTTASASATLAVYY755 pKa = 10.19GGRR758 pKa = 11.84FFAARR763 pKa = 11.84DD764 pKa = 3.48SDD766 pKa = 3.59RR767 pKa = 11.84TLQVLDD773 pKa = 4.36PATGTWTQGGAFTAGATAGRR793 pKa = 11.84LVRR796 pKa = 11.84MAIGPDD802 pKa = 3.3GTGYY806 pKa = 11.56AMDD809 pKa = 4.57GAGSFYY815 pKa = 10.53TFSTTAPYY823 pKa = 10.13PVTSGPRR830 pKa = 11.84TLTITSAGAPAFGGSGDD847 pKa = 4.54FIVDD851 pKa = 3.47SLGRR855 pKa = 11.84LYY857 pKa = 11.17LLSTAANSTYY867 pKa = 10.71VDD869 pKa = 4.34LYY871 pKa = 11.03EE872 pKa = 5.15LFASTSTAQYY882 pKa = 10.45LGRR885 pKa = 11.84ITNATVINTVFGGFAATPNGIFGRR909 pKa = 11.84SGTGRR914 pKa = 11.84LIQVDD919 pKa = 4.49LANLTVTQIGTDD931 pKa = 3.62VTPGSTDD938 pKa = 3.4LASCTYY944 pKa = 8.44PTFTRR949 pKa = 11.84TVTATQTVTKK959 pKa = 10.23VAGSTGTDD967 pKa = 3.25VRR969 pKa = 11.84PGDD972 pKa = 3.65TLEE975 pKa = 4.02YY976 pKa = 9.76TVVIRR981 pKa = 11.84NSGNIAAGQTTFQEE995 pKa = 4.9AIPAGTTYY1003 pKa = 10.8VPGTTTLNGTAVTDD1017 pKa = 3.84AGGAMPFTTAGSVNTPGQTAGVLSPDD1043 pKa = 3.59TTPGVLDD1050 pKa = 3.8RR1051 pKa = 11.84EE1052 pKa = 4.34AVVRR1056 pKa = 11.84FRR1058 pKa = 11.84VVVGTGTTSVSAQGTTTYY1076 pKa = 11.18SDD1078 pKa = 3.13VGTQTTLTDD1087 pKa = 3.88DD1088 pKa = 4.67PGTATANDD1096 pKa = 4.2PNVTPVNQPPVANNDD1111 pKa = 3.63AASTPAGSPVTFSVTANDD1129 pKa = 3.59TDD1131 pKa = 4.09TAPGTVNAATVDD1143 pKa = 4.31LNPSTAALDD1152 pKa = 3.79TTFTVPNQGTFTVNTAGQVTFTPVAGFSGIVTRR1185 pKa = 11.84TYY1187 pKa = 10.13TVQDD1191 pKa = 3.53NQGFTSNAATITVTVTPRR1209 pKa = 11.84AVNDD1213 pKa = 3.87TASTAPSASVNVPVLNNDD1231 pKa = 2.88VGTGLQPGSVVFVNAPAGSTVTNGGKK1257 pKa = 7.92TLTNAQGTYY1266 pKa = 8.47QVQPDD1271 pKa = 3.9GSVTFTPALGFTGTTTPVSYY1291 pKa = 9.55TVTDD1295 pKa = 3.54AAGTISNPATLTITVSSITAPTATNDD1321 pKa = 3.53AASTPKK1327 pKa = 9.38GTSVTLPGATNDD1339 pKa = 3.79TPGTLPIAPATVDD1352 pKa = 4.74LDD1354 pKa = 4.05PGTAGQQTTLSVTGGTFTANANGTVTFAPAAGFAGTATATYY1395 pKa = 9.03TVADD1399 pKa = 3.76TAGNRR1404 pKa = 11.84SNAATLTVTVTNQTPTATAATNATLPASAGPTALTPGLSGTDD1446 pKa = 3.3PDD1448 pKa = 4.03GTITTFTVTTLPPAGSGTLACGGTPITTVPTSCAPGQLTFDD1489 pKa = 4.08PAATFSGNAAFQFTVTDD1506 pKa = 4.13DD1507 pKa = 3.81NGATSAPATYY1517 pKa = 8.73TIPVDD1522 pKa = 4.01APPVAQNDD1530 pKa = 3.92VASTNPGVPVTFSVTANDD1548 pKa = 3.59TDD1550 pKa = 4.13TAPGTVDD1557 pKa = 3.13PASVVFVNPPAGSTLSPDD1575 pKa = 3.65GKK1577 pKa = 8.99TLTTPGVGTLSANADD1592 pKa = 3.5GTVTFTPAAGFTSGTLTTGYY1612 pKa = 7.95TVRR1615 pKa = 11.84DD1616 pKa = 3.51NLGQVSAPATITVSVPANTDD1636 pKa = 2.9VALSLTGPAFASPGQPVTYY1655 pKa = 9.57TLVVTNGGTNVTGTTVTVPLPAGTTFVSASAGGSVSGGTVTWTLGALAPTEE1706 pKa = 4.26TRR1708 pKa = 11.84TLTLTLTAPNATFINANGVTTLTTTGTVSVAQSEE1742 pKa = 4.85STTANNTDD1750 pKa = 3.1SSSAQLVYY1758 pKa = 10.84AQLTKK1763 pKa = 9.96QVRR1766 pKa = 11.84NVTRR1770 pKa = 11.84AGPFGVTATGQPNDD1784 pKa = 3.47VLEE1787 pKa = 4.12YY1788 pKa = 11.03CIDD1791 pKa = 3.52FRR1793 pKa = 11.84NAGGAALPNFTVLDD1807 pKa = 4.04PVPTGSAALPGAYY1820 pKa = 9.46DD1821 pKa = 3.58AAAGGAGFGVQLTRR1835 pKa = 11.84GGTTSYY1841 pKa = 9.53LTSFADD1847 pKa = 3.8ADD1849 pKa = 3.89NGQLDD1854 pKa = 3.78AAGLNVALGTLAVGEE1869 pKa = 4.19AGRR1872 pKa = 11.84TCFQARR1878 pKa = 11.84IPP1880 pKa = 3.85
MM1 pKa = 7.41HH2 pKa = 7.73PATRR6 pKa = 11.84TTPALIRR13 pKa = 11.84TLLTLAALLGLTWAPAHH30 pKa = 6.33AVTFNLSFAGGVDD43 pKa = 3.41VTTGSTCATALNSRR57 pKa = 11.84CRR59 pKa = 11.84FNSVVVGAGTGPYY72 pKa = 9.79QRR74 pKa = 11.84DD75 pKa = 3.56AIITISKK82 pKa = 7.98LTNAAINNSSVAAPNSDD99 pKa = 4.16LDD101 pKa = 3.98NPAPTLTGTAPAASAQDD118 pKa = 3.83FFSPTVDD125 pKa = 3.04VSSAGTGTGWAEE137 pKa = 3.53FTIDD141 pKa = 4.17FIARR145 pKa = 11.84GAAVPAVGAGNAALPGTFHH164 pKa = 5.85VTSFDD169 pKa = 3.32TDD171 pKa = 3.07AANPLRR177 pKa = 11.84EE178 pKa = 4.07FVEE181 pKa = 4.75YY182 pKa = 10.39VAPATTALSSGTTLTSRR199 pKa = 11.84AAVDD203 pKa = 3.27GGVAYY208 pKa = 10.53QSGTTGVSGISTDD221 pKa = 4.26PIYY224 pKa = 10.62KK225 pKa = 10.53ASATYY230 pKa = 10.17TNAASVRR237 pKa = 11.84LVYY240 pKa = 10.18GANTGGTACSGATCEE255 pKa = 4.67RR256 pKa = 11.84LTAFDD261 pKa = 5.35FYY263 pKa = 11.55VADD266 pKa = 3.98SVVLQTDD273 pKa = 3.48VNGFKK278 pKa = 10.48SARR281 pKa = 11.84LTTDD285 pKa = 2.79ADD287 pKa = 3.98GNGAITPGDD296 pKa = 3.57TVTYY300 pKa = 9.0TVTYY304 pKa = 10.65VNTGNTASTGFQITDD319 pKa = 3.69ALPSGVTTTAGAQTVRR335 pKa = 11.84LNGTVTAAARR345 pKa = 11.84NTAYY349 pKa = 9.7TGTGANTTLLAAGQTLPANGTISVDD374 pKa = 2.95IPVVVGAVTSDD385 pKa = 3.15NTVLSNQASGSGSGATNVLSDD406 pKa = 3.96NVDD409 pKa = 2.93ATTAFPPSVTGATGFAAPPAGSVPQTQTAAVSPTTITVRR448 pKa = 11.84RR449 pKa = 11.84QPTLTLNKK457 pKa = 9.3TIAAPGRR464 pKa = 11.84VNTNDD469 pKa = 2.96QFTVQISSGGTTVASATTTGTNTTATTGAVTVASGTTYY507 pKa = 10.55TLSEE511 pKa = 4.1ALAAGSGSTLSAYY524 pKa = 10.33ASTMTCTNATGGSSTTLPGGAGVSFTVTPTFGDD557 pKa = 3.86DD558 pKa = 3.59LTCTFTNTGTPVDD571 pKa = 4.15LAVSQTGPATANLDD585 pKa = 3.43GTVTYY590 pKa = 7.97TVRR593 pKa = 11.84VWNNGSDD600 pKa = 3.64VTGATFTDD608 pKa = 4.14TVPAALTNVTWTCAATGSASCGATTSGTGNTISATLGALTTDD650 pKa = 3.42TGSAATADD658 pKa = 3.77TNYY661 pKa = 8.86VTFTITGTATSTGSLTNTATVTAPGGFTEE690 pKa = 5.85SSTTNNSASTSTTVTATVQCSTLYY714 pKa = 9.53GTFGTGRR721 pKa = 11.84EE722 pKa = 4.12LRR724 pKa = 11.84SVTEE728 pKa = 4.12TNTVGSLIGTIPDD741 pKa = 3.21QGTTASASATLAVYY755 pKa = 10.19GGRR758 pKa = 11.84FFAARR763 pKa = 11.84DD764 pKa = 3.48SDD766 pKa = 3.59RR767 pKa = 11.84TLQVLDD773 pKa = 4.36PATGTWTQGGAFTAGATAGRR793 pKa = 11.84LVRR796 pKa = 11.84MAIGPDD802 pKa = 3.3GTGYY806 pKa = 11.56AMDD809 pKa = 4.57GAGSFYY815 pKa = 10.53TFSTTAPYY823 pKa = 10.13PVTSGPRR830 pKa = 11.84TLTITSAGAPAFGGSGDD847 pKa = 4.54FIVDD851 pKa = 3.47SLGRR855 pKa = 11.84LYY857 pKa = 11.17LLSTAANSTYY867 pKa = 10.71VDD869 pKa = 4.34LYY871 pKa = 11.03EE872 pKa = 5.15LFASTSTAQYY882 pKa = 10.45LGRR885 pKa = 11.84ITNATVINTVFGGFAATPNGIFGRR909 pKa = 11.84SGTGRR914 pKa = 11.84LIQVDD919 pKa = 4.49LANLTVTQIGTDD931 pKa = 3.62VTPGSTDD938 pKa = 3.4LASCTYY944 pKa = 8.44PTFTRR949 pKa = 11.84TVTATQTVTKK959 pKa = 10.23VAGSTGTDD967 pKa = 3.25VRR969 pKa = 11.84PGDD972 pKa = 3.65TLEE975 pKa = 4.02YY976 pKa = 9.76TVVIRR981 pKa = 11.84NSGNIAAGQTTFQEE995 pKa = 4.9AIPAGTTYY1003 pKa = 10.8VPGTTTLNGTAVTDD1017 pKa = 3.84AGGAMPFTTAGSVNTPGQTAGVLSPDD1043 pKa = 3.59TTPGVLDD1050 pKa = 3.8RR1051 pKa = 11.84EE1052 pKa = 4.34AVVRR1056 pKa = 11.84FRR1058 pKa = 11.84VVVGTGTTSVSAQGTTTYY1076 pKa = 11.18SDD1078 pKa = 3.13VGTQTTLTDD1087 pKa = 3.88DD1088 pKa = 4.67PGTATANDD1096 pKa = 4.2PNVTPVNQPPVANNDD1111 pKa = 3.63AASTPAGSPVTFSVTANDD1129 pKa = 3.59TDD1131 pKa = 4.09TAPGTVNAATVDD1143 pKa = 4.31LNPSTAALDD1152 pKa = 3.79TTFTVPNQGTFTVNTAGQVTFTPVAGFSGIVTRR1185 pKa = 11.84TYY1187 pKa = 10.13TVQDD1191 pKa = 3.53NQGFTSNAATITVTVTPRR1209 pKa = 11.84AVNDD1213 pKa = 3.87TASTAPSASVNVPVLNNDD1231 pKa = 2.88VGTGLQPGSVVFVNAPAGSTVTNGGKK1257 pKa = 7.92TLTNAQGTYY1266 pKa = 8.47QVQPDD1271 pKa = 3.9GSVTFTPALGFTGTTTPVSYY1291 pKa = 9.55TVTDD1295 pKa = 3.54AAGTISNPATLTITVSSITAPTATNDD1321 pKa = 3.53AASTPKK1327 pKa = 9.38GTSVTLPGATNDD1339 pKa = 3.79TPGTLPIAPATVDD1352 pKa = 4.74LDD1354 pKa = 4.05PGTAGQQTTLSVTGGTFTANANGTVTFAPAAGFAGTATATYY1395 pKa = 9.03TVADD1399 pKa = 3.76TAGNRR1404 pKa = 11.84SNAATLTVTVTNQTPTATAATNATLPASAGPTALTPGLSGTDD1446 pKa = 3.3PDD1448 pKa = 4.03GTITTFTVTTLPPAGSGTLACGGTPITTVPTSCAPGQLTFDD1489 pKa = 4.08PAATFSGNAAFQFTVTDD1506 pKa = 4.13DD1507 pKa = 3.81NGATSAPATYY1517 pKa = 8.73TIPVDD1522 pKa = 4.01APPVAQNDD1530 pKa = 3.92VASTNPGVPVTFSVTANDD1548 pKa = 3.59TDD1550 pKa = 4.13TAPGTVDD1557 pKa = 3.13PASVVFVNPPAGSTLSPDD1575 pKa = 3.65GKK1577 pKa = 8.99TLTTPGVGTLSANADD1592 pKa = 3.5GTVTFTPAAGFTSGTLTTGYY1612 pKa = 7.95TVRR1615 pKa = 11.84DD1616 pKa = 3.51NLGQVSAPATITVSVPANTDD1636 pKa = 2.9VALSLTGPAFASPGQPVTYY1655 pKa = 9.57TLVVTNGGTNVTGTTVTVPLPAGTTFVSASAGGSVSGGTVTWTLGALAPTEE1706 pKa = 4.26TRR1708 pKa = 11.84TLTLTLTAPNATFINANGVTTLTTTGTVSVAQSEE1742 pKa = 4.85STTANNTDD1750 pKa = 3.1SSSAQLVYY1758 pKa = 10.84AQLTKK1763 pKa = 9.96QVRR1766 pKa = 11.84NVTRR1770 pKa = 11.84AGPFGVTATGQPNDD1784 pKa = 3.47VLEE1787 pKa = 4.12YY1788 pKa = 11.03CIDD1791 pKa = 3.52FRR1793 pKa = 11.84NAGGAALPNFTVLDD1807 pKa = 4.04PVPTGSAALPGAYY1820 pKa = 9.46DD1821 pKa = 3.58AAAGGAGFGVQLTRR1835 pKa = 11.84GGTTSYY1841 pKa = 9.53LTSFADD1847 pKa = 3.8ADD1849 pKa = 3.89NGQLDD1854 pKa = 3.78AAGLNVALGTLAVGEE1869 pKa = 4.19AGRR1872 pKa = 11.84TCFQARR1878 pKa = 11.84IPP1880 pKa = 3.85
Molecular weight: 185.73 kDa
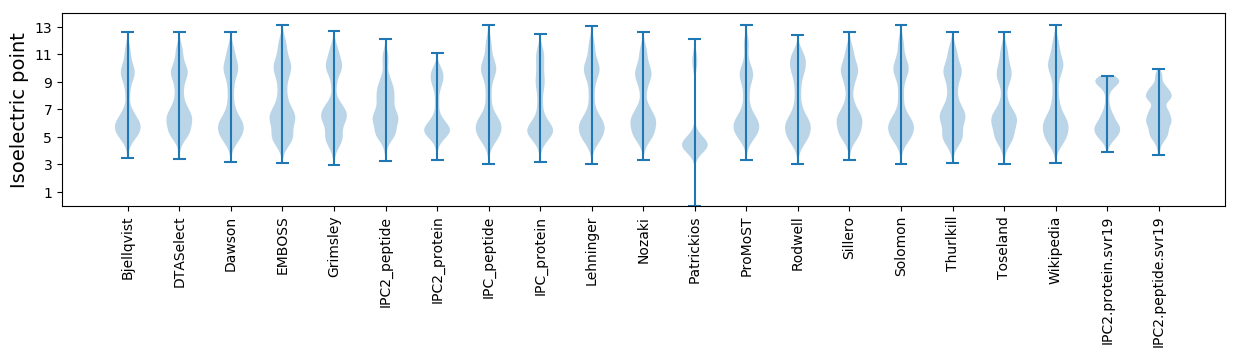
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E8U6Q2|E8U6Q2_DEIML Uncharacterized protein OS=Deinococcus maricopensis (strain DSM 21211 / LMG 22137 / NRRL B-23946 / LB-34) OX=709986 GN=Deima_1088 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.11QPNVRR10 pKa = 11.84KK11 pKa = 9.49RR12 pKa = 11.84AKK14 pKa = 8.27THH16 pKa = 5.41GFRR19 pKa = 11.84SRR21 pKa = 11.84MKK23 pKa = 9.3TKK25 pKa = 10.49AGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84HH40 pKa = 5.4RR41 pKa = 11.84LTVSVV46 pKa = 3.92
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 10.11QPNVRR10 pKa = 11.84KK11 pKa = 9.49RR12 pKa = 11.84AKK14 pKa = 8.27THH16 pKa = 5.41GFRR19 pKa = 11.84SRR21 pKa = 11.84MKK23 pKa = 9.3TKK25 pKa = 10.49AGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.03GRR39 pKa = 11.84HH40 pKa = 5.4RR41 pKa = 11.84LTVSVV46 pKa = 3.92
Molecular weight: 5.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1032088 |
30 |
3180 |
316.4 |
34.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.399 ± 0.064 | 0.524 ± 0.01 |
5.658 ± 0.029 | 4.913 ± 0.045 |
3.079 ± 0.023 | 8.691 ± 0.042 |
2.553 ± 0.033 | 3.35 ± 0.037 |
2.163 ± 0.033 | 11.485 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.779 ± 0.018 | 2.559 ± 0.029 |
5.708 ± 0.032 | 3.473 ± 0.03 |
7.78 ± 0.047 | 4.424 ± 0.03 |
6.721 ± 0.054 | 8.091 ± 0.039 |
1.278 ± 0.019 | 2.372 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
