
Ancylostoma ceylanicum
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Strongylida; Ancylostomatoidea; Ancylostomatidae; Ancylostomatinae; Ancylostoma
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
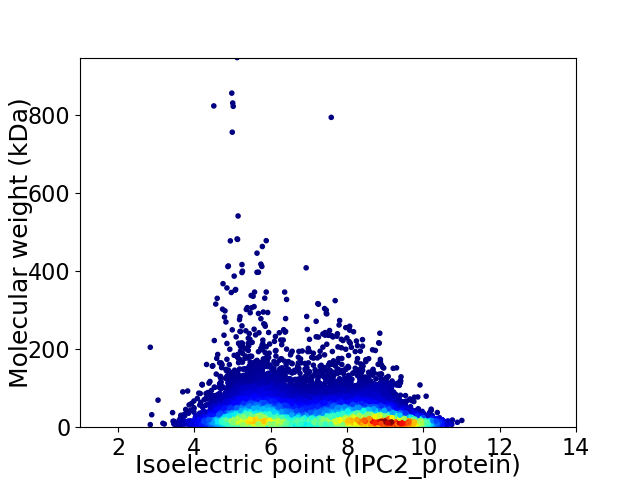
Virtual 2D-PAGE plot for 65375 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A016VAG5|A0A016VAG5_9BILA Isoform of A0A016VB49 EGF-like domain-containing protein OS=Ancylostoma ceylanicum OX=53326 GN=Acey_s0013.g2023 PE=4 SV=1
MM1 pKa = 7.48FSYY4 pKa = 10.34LYY6 pKa = 10.29KK7 pKa = 10.63VQLHH11 pKa = 5.64LHH13 pKa = 5.36TRR15 pKa = 11.84SFYY18 pKa = 10.47IGFPEE23 pKa = 3.77NTILFDD29 pKa = 4.25FSTPAFEE36 pKa = 4.79EE37 pKa = 4.57STEE40 pKa = 4.19GTTPTEE46 pKa = 4.34SMTSLYY52 pKa = 7.98TTMMHH57 pKa = 6.82EE58 pKa = 4.41SSTMSSLEE66 pKa = 4.08TTTMSVTDD74 pKa = 4.51GEE76 pKa = 4.67SSPSLISSTTDD87 pKa = 2.9GATSIQDD94 pKa = 3.67LSTSSGTEE102 pKa = 3.46EE103 pKa = 3.88STATDD108 pKa = 4.01GSSTSVMVSSIEE120 pKa = 3.93TTPAMVSSMEE130 pKa = 4.27TTPFTQSEE138 pKa = 4.45QPSTLEE144 pKa = 4.17VGSEE148 pKa = 4.18TPTVEE153 pKa = 4.36ISSTSHH159 pKa = 6.51DD160 pKa = 3.74TFPYY164 pKa = 9.87PDD166 pKa = 3.92STTQSASTLEE176 pKa = 4.36EE177 pKa = 4.03DD178 pKa = 3.66TSPFDD183 pKa = 3.36VSTTVGVPGSTAAMATTEE201 pKa = 4.01EE202 pKa = 4.68SMGPTAEE209 pKa = 4.14EE210 pKa = 3.71QTMTSAQSTDD220 pKa = 3.06STPNEE225 pKa = 4.23DD226 pKa = 3.51TSSSNTPEE234 pKa = 4.16FTSSSSVSFASSTQSSDD251 pKa = 2.97TDD253 pKa = 3.76GVTSTVSMTTSYY265 pKa = 11.7DD266 pKa = 3.92DD267 pKa = 3.5IVSEE271 pKa = 4.55TTTSSGGIEE280 pKa = 4.3VTTDD284 pKa = 3.07PSTIEE289 pKa = 4.07SSGSTEE295 pKa = 3.68NAAGSTEE302 pKa = 3.76ASSEE306 pKa = 3.88YY307 pKa = 10.58TEE309 pKa = 4.75FVTGSSVTVAQTTPDD324 pKa = 3.39STSIPSEE331 pKa = 4.26SPEE334 pKa = 4.02STTSSEE340 pKa = 4.0QSGLSSSISYY350 pKa = 7.67EE351 pKa = 4.2TTSEE355 pKa = 3.96AVTSSGHH362 pKa = 4.7TDD364 pKa = 3.31EE365 pKa = 5.63PGSTSSMGTTEE376 pKa = 4.58NVPSTDD382 pKa = 3.67GASSSSYY389 pKa = 10.11ISSSEE394 pKa = 3.95LTSSIASEE402 pKa = 4.12ATDD405 pKa = 4.04DD406 pKa = 4.2GAITEE411 pKa = 4.5STISPVVTSSVSGEE425 pKa = 4.25TTVGEE430 pKa = 4.14LTSAGVDD437 pKa = 3.71LSTSPSSDD445 pKa = 2.8GVSEE449 pKa = 4.39SSSSLSEE456 pKa = 4.08STTDD460 pKa = 3.44MAVTSSNPEE469 pKa = 3.79QPTDD473 pKa = 3.47TSITTVEE480 pKa = 4.12RR481 pKa = 11.84TSDD484 pKa = 3.44GVTASTDD491 pKa = 3.49VAVTSYY497 pKa = 11.4SEE499 pKa = 4.33GTSEE503 pKa = 4.16VPTVTGSGTPAEE515 pKa = 4.63STSSDD520 pKa = 3.48PTSAAVTSSGYY531 pKa = 9.75SEE533 pKa = 5.08EE534 pKa = 4.29PTTGHH539 pKa = 5.84STTVDD544 pKa = 3.2EE545 pKa = 5.09SGSTTAFFVTTGSDD559 pKa = 3.04LGASTTEE566 pKa = 3.7SSTAVMTSSEE576 pKa = 4.33FPEE579 pKa = 4.46VSTVPEE585 pKa = 3.89EE586 pKa = 4.35TITDD590 pKa = 4.44SITSEE595 pKa = 4.21SLPSEE600 pKa = 3.95NSVSQMSTTISHH612 pKa = 6.79TSSSAVSEE620 pKa = 4.21YY621 pKa = 10.62TDD623 pKa = 5.17LISTEE628 pKa = 3.95ISQDD632 pKa = 3.32TTTLPPWNIPSSTTDD647 pKa = 3.41DD648 pKa = 4.02TPTTTASWTSTTTISPTTTTSRR670 pKa = 11.84IPDD673 pKa = 4.45IITTTTTTAMPTVSSTLPRR692 pKa = 11.84TSTTTTSPTSTTTKK706 pKa = 10.24IPDD709 pKa = 4.23TITTTTATATTTTTTAMPTVPSTPPRR735 pKa = 11.84TSTTTTSPTSTTTKK749 pKa = 10.22IPDD752 pKa = 3.06ITATTTRR759 pKa = 11.84ATTTTTTTTITTTTAVPTISSTLLSTATVPTTTTVPTVTTGPSVCVDD806 pKa = 3.83GYY808 pKa = 11.29RR809 pKa = 11.84LHH811 pKa = 6.13QANAVVSSAGPGKK824 pKa = 10.46FGGFFTYY831 pKa = 8.73ITSCAEE837 pKa = 3.64LCEE840 pKa = 4.05RR841 pKa = 11.84DD842 pKa = 3.74YY843 pKa = 11.91GLSKK847 pKa = 10.92CLGFAYY853 pKa = 10.09EE854 pKa = 3.83PTNRR858 pKa = 11.84GKK860 pKa = 9.15CTLYY864 pKa = 10.19QRR866 pKa = 11.84SIGAIKK872 pKa = 9.52TDD874 pKa = 3.35SGSTASIYY882 pKa = 10.2KK883 pKa = 10.05RR884 pKa = 11.84CC885 pKa = 3.65
MM1 pKa = 7.48FSYY4 pKa = 10.34LYY6 pKa = 10.29KK7 pKa = 10.63VQLHH11 pKa = 5.64LHH13 pKa = 5.36TRR15 pKa = 11.84SFYY18 pKa = 10.47IGFPEE23 pKa = 3.77NTILFDD29 pKa = 4.25FSTPAFEE36 pKa = 4.79EE37 pKa = 4.57STEE40 pKa = 4.19GTTPTEE46 pKa = 4.34SMTSLYY52 pKa = 7.98TTMMHH57 pKa = 6.82EE58 pKa = 4.41SSTMSSLEE66 pKa = 4.08TTTMSVTDD74 pKa = 4.51GEE76 pKa = 4.67SSPSLISSTTDD87 pKa = 2.9GATSIQDD94 pKa = 3.67LSTSSGTEE102 pKa = 3.46EE103 pKa = 3.88STATDD108 pKa = 4.01GSSTSVMVSSIEE120 pKa = 3.93TTPAMVSSMEE130 pKa = 4.27TTPFTQSEE138 pKa = 4.45QPSTLEE144 pKa = 4.17VGSEE148 pKa = 4.18TPTVEE153 pKa = 4.36ISSTSHH159 pKa = 6.51DD160 pKa = 3.74TFPYY164 pKa = 9.87PDD166 pKa = 3.92STTQSASTLEE176 pKa = 4.36EE177 pKa = 4.03DD178 pKa = 3.66TSPFDD183 pKa = 3.36VSTTVGVPGSTAAMATTEE201 pKa = 4.01EE202 pKa = 4.68SMGPTAEE209 pKa = 4.14EE210 pKa = 3.71QTMTSAQSTDD220 pKa = 3.06STPNEE225 pKa = 4.23DD226 pKa = 3.51TSSSNTPEE234 pKa = 4.16FTSSSSVSFASSTQSSDD251 pKa = 2.97TDD253 pKa = 3.76GVTSTVSMTTSYY265 pKa = 11.7DD266 pKa = 3.92DD267 pKa = 3.5IVSEE271 pKa = 4.55TTTSSGGIEE280 pKa = 4.3VTTDD284 pKa = 3.07PSTIEE289 pKa = 4.07SSGSTEE295 pKa = 3.68NAAGSTEE302 pKa = 3.76ASSEE306 pKa = 3.88YY307 pKa = 10.58TEE309 pKa = 4.75FVTGSSVTVAQTTPDD324 pKa = 3.39STSIPSEE331 pKa = 4.26SPEE334 pKa = 4.02STTSSEE340 pKa = 4.0QSGLSSSISYY350 pKa = 7.67EE351 pKa = 4.2TTSEE355 pKa = 3.96AVTSSGHH362 pKa = 4.7TDD364 pKa = 3.31EE365 pKa = 5.63PGSTSSMGTTEE376 pKa = 4.58NVPSTDD382 pKa = 3.67GASSSSYY389 pKa = 10.11ISSSEE394 pKa = 3.95LTSSIASEE402 pKa = 4.12ATDD405 pKa = 4.04DD406 pKa = 4.2GAITEE411 pKa = 4.5STISPVVTSSVSGEE425 pKa = 4.25TTVGEE430 pKa = 4.14LTSAGVDD437 pKa = 3.71LSTSPSSDD445 pKa = 2.8GVSEE449 pKa = 4.39SSSSLSEE456 pKa = 4.08STTDD460 pKa = 3.44MAVTSSNPEE469 pKa = 3.79QPTDD473 pKa = 3.47TSITTVEE480 pKa = 4.12RR481 pKa = 11.84TSDD484 pKa = 3.44GVTASTDD491 pKa = 3.49VAVTSYY497 pKa = 11.4SEE499 pKa = 4.33GTSEE503 pKa = 4.16VPTVTGSGTPAEE515 pKa = 4.63STSSDD520 pKa = 3.48PTSAAVTSSGYY531 pKa = 9.75SEE533 pKa = 5.08EE534 pKa = 4.29PTTGHH539 pKa = 5.84STTVDD544 pKa = 3.2EE545 pKa = 5.09SGSTTAFFVTTGSDD559 pKa = 3.04LGASTTEE566 pKa = 3.7SSTAVMTSSEE576 pKa = 4.33FPEE579 pKa = 4.46VSTVPEE585 pKa = 3.89EE586 pKa = 4.35TITDD590 pKa = 4.44SITSEE595 pKa = 4.21SLPSEE600 pKa = 3.95NSVSQMSTTISHH612 pKa = 6.79TSSSAVSEE620 pKa = 4.21YY621 pKa = 10.62TDD623 pKa = 5.17LISTEE628 pKa = 3.95ISQDD632 pKa = 3.32TTTLPPWNIPSSTTDD647 pKa = 3.41DD648 pKa = 4.02TPTTTASWTSTTTISPTTTTSRR670 pKa = 11.84IPDD673 pKa = 4.45IITTTTTTAMPTVSSTLPRR692 pKa = 11.84TSTTTTSPTSTTTKK706 pKa = 10.24IPDD709 pKa = 4.23TITTTTATATTTTTTAMPTVPSTPPRR735 pKa = 11.84TSTTTTSPTSTTTKK749 pKa = 10.22IPDD752 pKa = 3.06ITATTTRR759 pKa = 11.84ATTTTTTTTITTTTAVPTISSTLLSTATVPTTTTVPTVTTGPSVCVDD806 pKa = 3.83GYY808 pKa = 11.29RR809 pKa = 11.84LHH811 pKa = 6.13QANAVVSSAGPGKK824 pKa = 10.46FGGFFTYY831 pKa = 8.73ITSCAEE837 pKa = 3.64LCEE840 pKa = 4.05RR841 pKa = 11.84DD842 pKa = 3.74YY843 pKa = 11.91GLSKK847 pKa = 10.92CLGFAYY853 pKa = 10.09EE854 pKa = 3.83PTNRR858 pKa = 11.84GKK860 pKa = 9.15CTLYY864 pKa = 10.19QRR866 pKa = 11.84SIGAIKK872 pKa = 9.52TDD874 pKa = 3.35SGSTASIYY882 pKa = 10.2KK883 pKa = 10.05RR884 pKa = 11.84CC885 pKa = 3.65
Molecular weight: 90.71 kDa
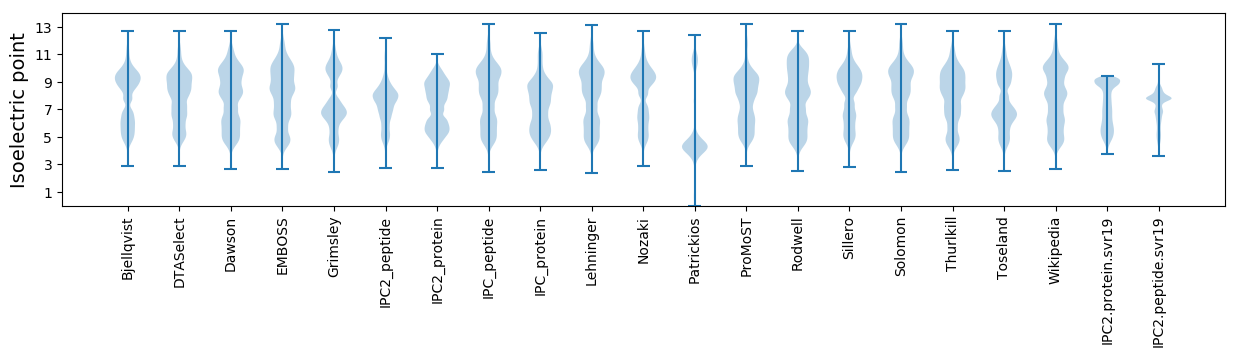
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A016VEV8|A0A016VEV8_9BILA Uncharacterized protein OS=Ancylostoma ceylanicum OX=53326 GN=Acey_s0011.g1590 PE=3 SV=1
MM1 pKa = 7.07HH2 pKa = 6.52QQFIALRR9 pKa = 11.84RR10 pKa = 11.84SLPVPLFSIIFLVLVIVAVILLISIIPALIGCLTMKK46 pKa = 10.28SKK48 pKa = 10.79RR49 pKa = 11.84NKK51 pKa = 8.9KK52 pKa = 8.81RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84SGKK58 pKa = 9.21HH59 pKa = 4.97SSSTMSGKK67 pKa = 9.91SSKK70 pKa = 10.41RR71 pKa = 11.84SGKK74 pKa = 10.62GKK76 pKa = 10.33GGPNRR81 pKa = 11.84KK82 pKa = 9.41RR83 pKa = 11.84GGKK86 pKa = 8.43SKK88 pKa = 10.6KK89 pKa = 10.03SSTSTTSTKK98 pKa = 10.18TGTSTRR104 pKa = 11.84SGKK107 pKa = 10.33KK108 pKa = 9.33SASKK112 pKa = 10.6SKK114 pKa = 10.58SSQLSEE120 pKa = 3.87KK121 pKa = 9.85SARR124 pKa = 11.84GAGRR128 pKa = 11.84NGRR131 pKa = 11.84PANKK135 pKa = 9.82GMKK138 pKa = 9.62SKK140 pKa = 10.75SKK142 pKa = 10.65SKK144 pKa = 10.66SKK146 pKa = 10.78SAGKK150 pKa = 9.41SGQMRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84KK158 pKa = 9.51SRR160 pKa = 11.84RR161 pKa = 3.25
MM1 pKa = 7.07HH2 pKa = 6.52QQFIALRR9 pKa = 11.84RR10 pKa = 11.84SLPVPLFSIIFLVLVIVAVILLISIIPALIGCLTMKK46 pKa = 10.28SKK48 pKa = 10.79RR49 pKa = 11.84NKK51 pKa = 8.9KK52 pKa = 8.81RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84SGKK58 pKa = 9.21HH59 pKa = 4.97SSSTMSGKK67 pKa = 9.91SSKK70 pKa = 10.41RR71 pKa = 11.84SGKK74 pKa = 10.62GKK76 pKa = 10.33GGPNRR81 pKa = 11.84KK82 pKa = 9.41RR83 pKa = 11.84GGKK86 pKa = 8.43SKK88 pKa = 10.6KK89 pKa = 10.03SSTSTTSTKK98 pKa = 10.18TGTSTRR104 pKa = 11.84SGKK107 pKa = 10.33KK108 pKa = 9.33SASKK112 pKa = 10.6SKK114 pKa = 10.58SSQLSEE120 pKa = 3.87KK121 pKa = 9.85SARR124 pKa = 11.84GAGRR128 pKa = 11.84NGRR131 pKa = 11.84PANKK135 pKa = 9.82GMKK138 pKa = 9.62SKK140 pKa = 10.75SKK142 pKa = 10.65SKK144 pKa = 10.66SKK146 pKa = 10.78SAGKK150 pKa = 9.41SGQMRR155 pKa = 11.84RR156 pKa = 11.84RR157 pKa = 11.84KK158 pKa = 9.51SRR160 pKa = 11.84RR161 pKa = 3.25
Molecular weight: 17.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
20039791 |
17 |
8404 |
306.5 |
34.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.207 ± 0.012 | 2.343 ± 0.011 |
5.199 ± 0.009 | 6.332 ± 0.016 |
4.103 ± 0.008 | 5.657 ± 0.013 |
2.467 ± 0.005 | 5.236 ± 0.01 |
5.556 ± 0.014 | 8.896 ± 0.014 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.006 | 4.08 ± 0.008 |
5.285 ± 0.013 | 3.803 ± 0.009 |
6.531 ± 0.013 | 8.016 ± 0.014 |
5.868 ± 0.015 | 6.641 ± 0.009 |
1.218 ± 0.004 | 3.004 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
