
Paracoccus aminovorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales;
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
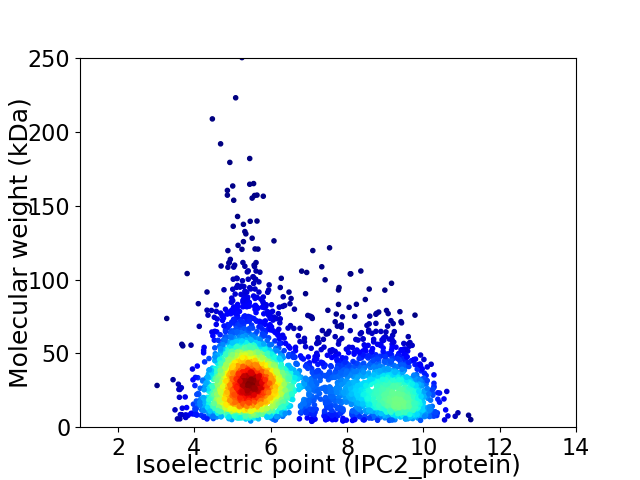
Virtual 2D-PAGE plot for 3736 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
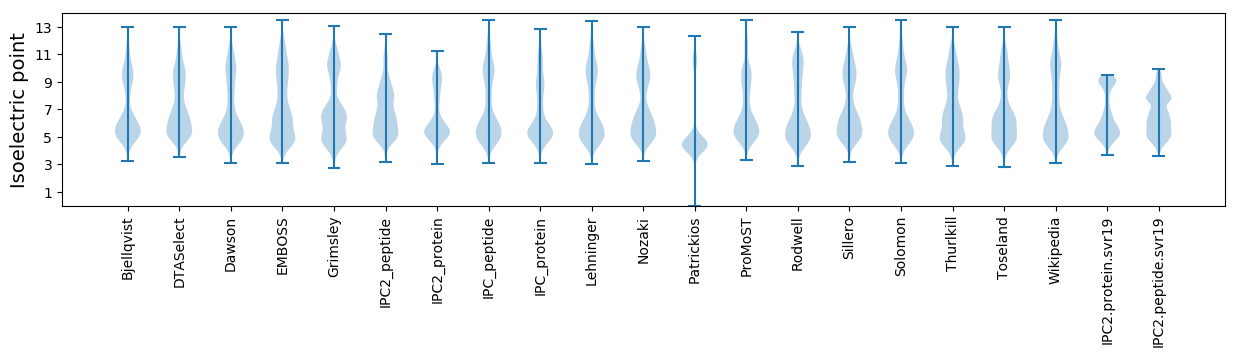
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3FJQ1|A0A1I3FJQ1_9RHOB PPC domain-containing protein OS=Paracoccus aminovorans OX=34004 GN=SAMN04488021_1794 PE=4 SV=1
MM1 pKa = 7.51ANAVGRR7 pKa = 11.84TFTAIGFLLEE17 pKa = 4.07SPEE20 pKa = 4.02IVEE23 pKa = 4.8ALHH26 pKa = 5.84TRR28 pKa = 11.84QGLAGVTTQFAGAIAGVAGGLWGASIGAAAAASMAAALVGLLAGMPITILSAVAVVGISFIAASISEE95 pKa = 5.01AIVEE99 pKa = 4.37SLVTQLFEE107 pKa = 5.42GMGDD111 pKa = 3.59DD112 pKa = 5.66LKK114 pKa = 11.7VHH116 pKa = 6.19TNDD119 pKa = 2.43VDD121 pKa = 4.0VYY123 pKa = 10.3IGGLRR128 pKa = 11.84GEE130 pKa = 4.31TLMAGSADD138 pKa = 3.33DD139 pKa = 3.75FVYY142 pKa = 10.83GRR144 pKa = 11.84GGNDD148 pKa = 3.34NISGGTGSDD157 pKa = 3.31HH158 pKa = 7.33LLGGSGNDD166 pKa = 3.78TITGGSGDD174 pKa = 4.25DD175 pKa = 4.28FISGGSGSNNMTGGVGNDD193 pKa = 3.19SYY195 pKa = 11.71VVEE198 pKa = 4.51SEE200 pKa = 3.78GDD202 pKa = 3.72TVNEE206 pKa = 4.04NVNGGVDD213 pKa = 3.6KK214 pKa = 10.84IVANISINLNDD225 pKa = 3.23ARR227 pKa = 11.84YY228 pKa = 10.19ANVEE232 pKa = 3.9NVTLIGEE239 pKa = 4.51SNISATGSDD248 pKa = 4.18HH249 pKa = 7.51DD250 pKa = 4.5NSLIGYY256 pKa = 8.51LGNNRR261 pKa = 11.84LYY263 pKa = 11.38GNDD266 pKa = 3.72GDD268 pKa = 4.53DD269 pKa = 4.43HH270 pKa = 8.24LSGSYY275 pKa = 11.32GNDD278 pKa = 2.88TLYY281 pKa = 11.3GGGGNDD287 pKa = 3.5TLNGGFGLDD296 pKa = 3.78SMIGGAGNDD305 pKa = 3.08IYY307 pKa = 11.62YY308 pKa = 10.4VDD310 pKa = 4.69DD311 pKa = 4.96LGDD314 pKa = 3.68VVIEE318 pKa = 4.15GSNGGEE324 pKa = 3.9DD325 pKa = 4.16TINSAISINLDD336 pKa = 3.46RR337 pKa = 11.84PNDD340 pKa = 3.27AYY342 pKa = 11.0ANVEE346 pKa = 4.14HH347 pKa = 6.84VVLQGTEE354 pKa = 3.98NINAYY359 pKa = 10.33GSVSDD364 pKa = 5.05NSLTGNRR371 pKa = 11.84GNNLLNGRR379 pKa = 11.84AGNDD383 pKa = 3.85SIVGAAGNDD392 pKa = 3.64TLYY395 pKa = 11.45GEE397 pKa = 4.97AGNDD401 pKa = 3.3ILNGGAGVDD410 pKa = 3.78SMVGGTGNDD419 pKa = 2.82IYY421 pKa = 11.02FVDD424 pKa = 5.55DD425 pKa = 3.2IRR427 pKa = 11.84DD428 pKa = 3.92IIVEE432 pKa = 4.25TATGGTDD439 pKa = 3.97RR440 pKa = 11.84INSSISIDD448 pKa = 3.64LNRR451 pKa = 11.84SGDD454 pKa = 3.46VYY456 pKa = 11.72ANIEE460 pKa = 4.16NIILLGAQNLNAYY473 pKa = 9.9GSGGSNNITGNSGANIISGRR493 pKa = 11.84YY494 pKa = 9.56GNDD497 pKa = 2.78NLAGGAGADD506 pKa = 3.2TFIFSKK512 pKa = 10.9NYY514 pKa = 9.5DD515 pKa = 3.25QDD517 pKa = 4.0VITDD521 pKa = 4.31FEE523 pKa = 5.95DD524 pKa = 3.35NTDD527 pKa = 4.06TIRR530 pKa = 11.84LLNFGVTTFTQAQGYY545 pKa = 8.45AADD548 pKa = 3.6WCKK551 pKa = 10.9CGII554 pKa = 4.05
MM1 pKa = 7.51ANAVGRR7 pKa = 11.84TFTAIGFLLEE17 pKa = 4.07SPEE20 pKa = 4.02IVEE23 pKa = 4.8ALHH26 pKa = 5.84TRR28 pKa = 11.84QGLAGVTTQFAGAIAGVAGGLWGASIGAAAAASMAAALVGLLAGMPITILSAVAVVGISFIAASISEE95 pKa = 5.01AIVEE99 pKa = 4.37SLVTQLFEE107 pKa = 5.42GMGDD111 pKa = 3.59DD112 pKa = 5.66LKK114 pKa = 11.7VHH116 pKa = 6.19TNDD119 pKa = 2.43VDD121 pKa = 4.0VYY123 pKa = 10.3IGGLRR128 pKa = 11.84GEE130 pKa = 4.31TLMAGSADD138 pKa = 3.33DD139 pKa = 3.75FVYY142 pKa = 10.83GRR144 pKa = 11.84GGNDD148 pKa = 3.34NISGGTGSDD157 pKa = 3.31HH158 pKa = 7.33LLGGSGNDD166 pKa = 3.78TITGGSGDD174 pKa = 4.25DD175 pKa = 4.28FISGGSGSNNMTGGVGNDD193 pKa = 3.19SYY195 pKa = 11.71VVEE198 pKa = 4.51SEE200 pKa = 3.78GDD202 pKa = 3.72TVNEE206 pKa = 4.04NVNGGVDD213 pKa = 3.6KK214 pKa = 10.84IVANISINLNDD225 pKa = 3.23ARR227 pKa = 11.84YY228 pKa = 10.19ANVEE232 pKa = 3.9NVTLIGEE239 pKa = 4.51SNISATGSDD248 pKa = 4.18HH249 pKa = 7.51DD250 pKa = 4.5NSLIGYY256 pKa = 8.51LGNNRR261 pKa = 11.84LYY263 pKa = 11.38GNDD266 pKa = 3.72GDD268 pKa = 4.53DD269 pKa = 4.43HH270 pKa = 8.24LSGSYY275 pKa = 11.32GNDD278 pKa = 2.88TLYY281 pKa = 11.3GGGGNDD287 pKa = 3.5TLNGGFGLDD296 pKa = 3.78SMIGGAGNDD305 pKa = 3.08IYY307 pKa = 11.62YY308 pKa = 10.4VDD310 pKa = 4.69DD311 pKa = 4.96LGDD314 pKa = 3.68VVIEE318 pKa = 4.15GSNGGEE324 pKa = 3.9DD325 pKa = 4.16TINSAISINLDD336 pKa = 3.46RR337 pKa = 11.84PNDD340 pKa = 3.27AYY342 pKa = 11.0ANVEE346 pKa = 4.14HH347 pKa = 6.84VVLQGTEE354 pKa = 3.98NINAYY359 pKa = 10.33GSVSDD364 pKa = 5.05NSLTGNRR371 pKa = 11.84GNNLLNGRR379 pKa = 11.84AGNDD383 pKa = 3.85SIVGAAGNDD392 pKa = 3.64TLYY395 pKa = 11.45GEE397 pKa = 4.97AGNDD401 pKa = 3.3ILNGGAGVDD410 pKa = 3.78SMVGGTGNDD419 pKa = 2.82IYY421 pKa = 11.02FVDD424 pKa = 5.55DD425 pKa = 3.2IRR427 pKa = 11.84DD428 pKa = 3.92IIVEE432 pKa = 4.25TATGGTDD439 pKa = 3.97RR440 pKa = 11.84INSSISIDD448 pKa = 3.64LNRR451 pKa = 11.84SGDD454 pKa = 3.46VYY456 pKa = 11.72ANIEE460 pKa = 4.16NIILLGAQNLNAYY473 pKa = 9.9GSGGSNNITGNSGANIISGRR493 pKa = 11.84YY494 pKa = 9.56GNDD497 pKa = 2.78NLAGGAGADD506 pKa = 3.2TFIFSKK512 pKa = 10.9NYY514 pKa = 9.5DD515 pKa = 3.25QDD517 pKa = 4.0VITDD521 pKa = 4.31FEE523 pKa = 5.95DD524 pKa = 3.35NTDD527 pKa = 4.06TIRR530 pKa = 11.84LLNFGVTTFTQAQGYY545 pKa = 8.45AADD548 pKa = 3.6WCKK551 pKa = 10.9CGII554 pKa = 4.05
Molecular weight: 56.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3BQP5|A0A1I3BQP5_9RHOB Two-component system OmpR family response regulator OS=Paracoccus aminovorans OX=34004 GN=SAMN04488021_1245 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.12GGRR29 pKa = 11.84RR30 pKa = 11.84VLNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84KK41 pKa = 6.51TLSAA45 pKa = 4.15
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84ARR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.37GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.12GGRR29 pKa = 11.84RR30 pKa = 11.84VLNARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84KK41 pKa = 6.51TLSAA45 pKa = 4.15
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1156815 |
39 |
2363 |
309.6 |
33.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.476 ± 0.068 | 0.836 ± 0.01 |
5.795 ± 0.029 | 5.628 ± 0.034 |
3.453 ± 0.024 | 9.048 ± 0.038 |
2.033 ± 0.018 | 4.788 ± 0.029 |
2.636 ± 0.035 | 10.449 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.608 ± 0.018 | 2.293 ± 0.029 |
5.65 ± 0.031 | 3.269 ± 0.023 |
7.789 ± 0.047 | 4.781 ± 0.027 |
4.966 ± 0.027 | 6.993 ± 0.03 |
1.493 ± 0.017 | 2.016 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
