
Paenibacillus cellulosilyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
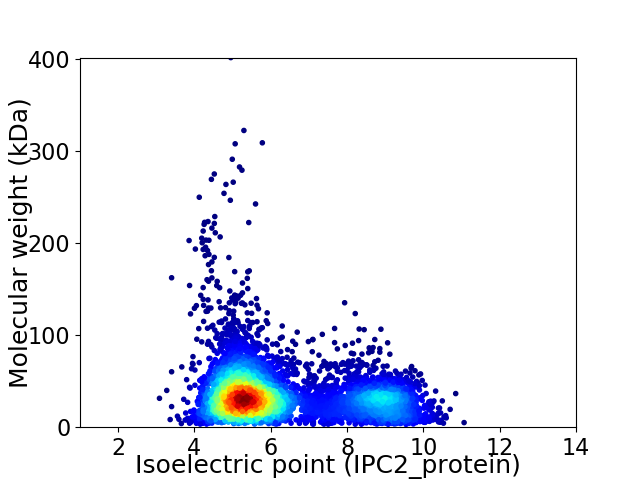
Virtual 2D-PAGE plot for 6112 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V2YLC8|A0A2V2YLC8_9BACL Uncharacterized protein OS=Paenibacillus cellulosilyticus OX=375489 GN=DFQ01_13147 PE=4 SV=1
MM1 pKa = 7.37TGSASYY7 pKa = 10.95SFSLHH12 pKa = 5.97NNGGGTVVVYY22 pKa = 9.5STMSLPVSDD31 pKa = 5.24PPSLDD36 pKa = 2.92HH37 pKa = 7.5DD38 pKa = 4.6AYY40 pKa = 10.71YY41 pKa = 10.11PIYY44 pKa = 9.61TRR46 pKa = 11.84LGTVDD51 pKa = 3.9SNQSGSFSTGNPLARR66 pKa = 11.84IVIARR71 pKa = 11.84ATDD74 pKa = 3.71DD75 pKa = 4.35FPLKK79 pKa = 10.5LAIVDD84 pKa = 3.93TLDD87 pKa = 4.27PDD89 pKa = 3.93SQTVAVGDD97 pKa = 4.18DD98 pKa = 3.87DD99 pKa = 5.24GKK101 pKa = 10.42ATALAWTFYY110 pKa = 10.8QSYY113 pKa = 10.62ASQPYY118 pKa = 9.38QPLALQFNDD127 pKa = 3.81LVLNNTDD134 pKa = 3.36PTQLNGQAASFFSANGYY151 pKa = 10.54AGIDD155 pKa = 3.34FSIFSMVSFWATNSLYY171 pKa = 10.66AFPGTYY177 pKa = 9.71YY178 pKa = 10.58CYY180 pKa = 10.21EE181 pKa = 4.35PQSDD185 pKa = 3.55THH187 pKa = 7.14GFIVPPDD194 pKa = 3.62EE195 pKa = 4.51VGTITVANGTATYY208 pKa = 9.72TSAADD213 pKa = 3.87GSTQTLLFVRR223 pKa = 11.84SKK225 pKa = 10.83LSSAGADD232 pKa = 3.48SKK234 pKa = 11.76NGIALTSILRR244 pKa = 11.84DD245 pKa = 3.6FAWEE249 pKa = 4.19GKK251 pKa = 8.95PDD253 pKa = 3.56QIGICFVGTNNGAQMIAQPYY273 pKa = 8.38QNPSVPWYY281 pKa = 10.03AVAYY285 pKa = 10.1DD286 pKa = 3.64MVYY289 pKa = 10.85GAFTLVQIAMALDD302 pKa = 3.81MAVHH306 pKa = 6.03MLKK309 pKa = 10.67SIGSGLQYY317 pKa = 11.15LKK319 pKa = 11.28DD320 pKa = 4.03NLGKK324 pKa = 10.7LYY326 pKa = 10.95DD327 pKa = 5.46NIRR330 pKa = 11.84QWANEE335 pKa = 4.24TGDD338 pKa = 3.61SAGPGSGIGDD348 pKa = 3.6DD349 pKa = 3.74VDD351 pKa = 3.89PVNVDD356 pKa = 2.56VDD358 pKa = 3.49IDD360 pKa = 3.7VDD362 pKa = 3.53IDD364 pKa = 3.44VDD366 pKa = 3.58IDD368 pKa = 3.46VDD370 pKa = 3.53IDD372 pKa = 3.17IDD374 pKa = 3.5IDD376 pKa = 3.52IDD378 pKa = 3.78VDD380 pKa = 3.55VDD382 pKa = 3.09IDD384 pKa = 3.42IDD386 pKa = 3.71IDD388 pKa = 3.77VDD390 pKa = 4.02FLAVVDD396 pKa = 3.93VDD398 pKa = 3.58VDD400 pKa = 3.53VDD402 pKa = 3.33IDD404 pKa = 3.43IDD406 pKa = 3.76IDD408 pKa = 4.04VVTDD412 pKa = 3.44TEE414 pKa = 4.34TDD416 pKa = 2.83VDD418 pKa = 3.67IDD420 pKa = 3.7VDD422 pKa = 3.63IDD424 pKa = 3.19IDD426 pKa = 3.85TDD428 pKa = 3.54VDD430 pKa = 3.92VQPGMIRR437 pKa = 11.84GLLNKK442 pKa = 9.54VGSWVMEE449 pKa = 4.07KK450 pKa = 10.6ALPALTEE457 pKa = 3.81MLAINLAFMSAAQLLQAWRR476 pKa = 11.84HH477 pKa = 5.37ADD479 pKa = 3.33EE480 pKa = 4.6QGIKK484 pKa = 10.2NLQPRR489 pKa = 11.84EE490 pKa = 4.16STGLGLLVNYY500 pKa = 7.63MLNEE504 pKa = 3.93QVSLQTRR511 pKa = 11.84WNTFADD517 pKa = 4.19YY518 pKa = 10.46VQQSQADD525 pKa = 3.55NATIEE530 pKa = 4.48MTVSTIIMAGNSAADD545 pKa = 3.43QAASNWRR552 pKa = 11.84WSTDD556 pKa = 3.04AEE558 pKa = 4.27NAAVAALAQYY568 pKa = 9.91TGDD571 pKa = 3.14NRR573 pKa = 11.84YY574 pKa = 8.73QAFQTLGGYY583 pKa = 9.14QYY585 pKa = 10.73QGRR588 pKa = 11.84DD589 pKa = 3.39LPVKK593 pKa = 10.57VGAQVAMDD601 pKa = 3.88YY602 pKa = 10.8LKK604 pKa = 11.04KK605 pKa = 10.87GGG607 pKa = 3.63
MM1 pKa = 7.37TGSASYY7 pKa = 10.95SFSLHH12 pKa = 5.97NNGGGTVVVYY22 pKa = 9.5STMSLPVSDD31 pKa = 5.24PPSLDD36 pKa = 2.92HH37 pKa = 7.5DD38 pKa = 4.6AYY40 pKa = 10.71YY41 pKa = 10.11PIYY44 pKa = 9.61TRR46 pKa = 11.84LGTVDD51 pKa = 3.9SNQSGSFSTGNPLARR66 pKa = 11.84IVIARR71 pKa = 11.84ATDD74 pKa = 3.71DD75 pKa = 4.35FPLKK79 pKa = 10.5LAIVDD84 pKa = 3.93TLDD87 pKa = 4.27PDD89 pKa = 3.93SQTVAVGDD97 pKa = 4.18DD98 pKa = 3.87DD99 pKa = 5.24GKK101 pKa = 10.42ATALAWTFYY110 pKa = 10.8QSYY113 pKa = 10.62ASQPYY118 pKa = 9.38QPLALQFNDD127 pKa = 3.81LVLNNTDD134 pKa = 3.36PTQLNGQAASFFSANGYY151 pKa = 10.54AGIDD155 pKa = 3.34FSIFSMVSFWATNSLYY171 pKa = 10.66AFPGTYY177 pKa = 9.71YY178 pKa = 10.58CYY180 pKa = 10.21EE181 pKa = 4.35PQSDD185 pKa = 3.55THH187 pKa = 7.14GFIVPPDD194 pKa = 3.62EE195 pKa = 4.51VGTITVANGTATYY208 pKa = 9.72TSAADD213 pKa = 3.87GSTQTLLFVRR223 pKa = 11.84SKK225 pKa = 10.83LSSAGADD232 pKa = 3.48SKK234 pKa = 11.76NGIALTSILRR244 pKa = 11.84DD245 pKa = 3.6FAWEE249 pKa = 4.19GKK251 pKa = 8.95PDD253 pKa = 3.56QIGICFVGTNNGAQMIAQPYY273 pKa = 8.38QNPSVPWYY281 pKa = 10.03AVAYY285 pKa = 10.1DD286 pKa = 3.64MVYY289 pKa = 10.85GAFTLVQIAMALDD302 pKa = 3.81MAVHH306 pKa = 6.03MLKK309 pKa = 10.67SIGSGLQYY317 pKa = 11.15LKK319 pKa = 11.28DD320 pKa = 4.03NLGKK324 pKa = 10.7LYY326 pKa = 10.95DD327 pKa = 5.46NIRR330 pKa = 11.84QWANEE335 pKa = 4.24TGDD338 pKa = 3.61SAGPGSGIGDD348 pKa = 3.6DD349 pKa = 3.74VDD351 pKa = 3.89PVNVDD356 pKa = 2.56VDD358 pKa = 3.49IDD360 pKa = 3.7VDD362 pKa = 3.53IDD364 pKa = 3.44VDD366 pKa = 3.58IDD368 pKa = 3.46VDD370 pKa = 3.53IDD372 pKa = 3.17IDD374 pKa = 3.5IDD376 pKa = 3.52IDD378 pKa = 3.78VDD380 pKa = 3.55VDD382 pKa = 3.09IDD384 pKa = 3.42IDD386 pKa = 3.71IDD388 pKa = 3.77VDD390 pKa = 4.02FLAVVDD396 pKa = 3.93VDD398 pKa = 3.58VDD400 pKa = 3.53VDD402 pKa = 3.33IDD404 pKa = 3.43IDD406 pKa = 3.76IDD408 pKa = 4.04VVTDD412 pKa = 3.44TEE414 pKa = 4.34TDD416 pKa = 2.83VDD418 pKa = 3.67IDD420 pKa = 3.7VDD422 pKa = 3.63IDD424 pKa = 3.19IDD426 pKa = 3.85TDD428 pKa = 3.54VDD430 pKa = 3.92VQPGMIRR437 pKa = 11.84GLLNKK442 pKa = 9.54VGSWVMEE449 pKa = 4.07KK450 pKa = 10.6ALPALTEE457 pKa = 3.81MLAINLAFMSAAQLLQAWRR476 pKa = 11.84HH477 pKa = 5.37ADD479 pKa = 3.33EE480 pKa = 4.6QGIKK484 pKa = 10.2NLQPRR489 pKa = 11.84EE490 pKa = 4.16STGLGLLVNYY500 pKa = 7.63MLNEE504 pKa = 3.93QVSLQTRR511 pKa = 11.84WNTFADD517 pKa = 4.19YY518 pKa = 10.46VQQSQADD525 pKa = 3.55NATIEE530 pKa = 4.48MTVSTIIMAGNSAADD545 pKa = 3.43QAASNWRR552 pKa = 11.84WSTDD556 pKa = 3.04AEE558 pKa = 4.27NAAVAALAQYY568 pKa = 9.91TGDD571 pKa = 3.14NRR573 pKa = 11.84YY574 pKa = 8.73QAFQTLGGYY583 pKa = 9.14QYY585 pKa = 10.73QGRR588 pKa = 11.84DD589 pKa = 3.39LPVKK593 pKa = 10.57VGAQVAMDD601 pKa = 3.88YY602 pKa = 10.8LKK604 pKa = 11.04KK605 pKa = 10.87GGG607 pKa = 3.63
Molecular weight: 65.57 kDa
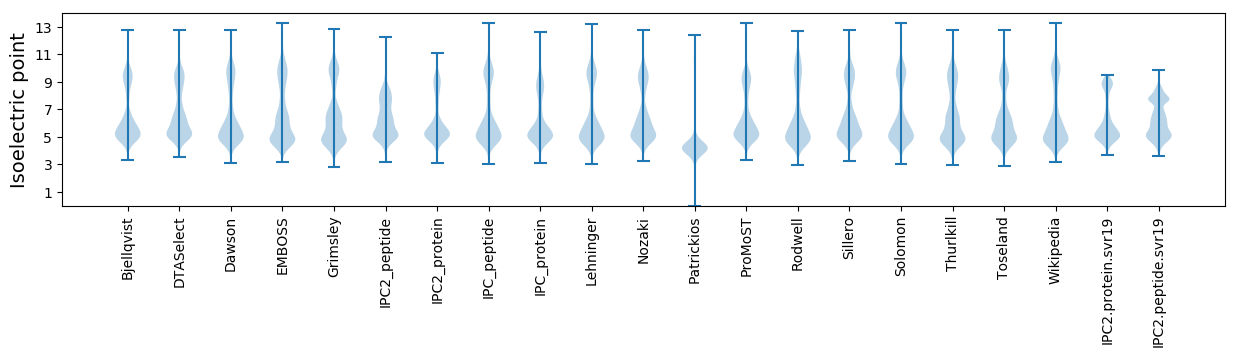
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V2Z1F9|A0A2V2Z1F9_9BACL Uncharacterized protein OS=Paenibacillus cellulosilyticus OX=375489 GN=DFQ01_11095 PE=4 SV=1
MM1 pKa = 7.61GPTFKK6 pKa = 10.87PNVSKK11 pKa = 10.88RR12 pKa = 11.84KK13 pKa = 8.93KK14 pKa = 8.63NHH16 pKa = 4.89GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MASKK25 pKa = 10.64NGRR28 pKa = 11.84KK29 pKa = 9.12VLAARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.23GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
MM1 pKa = 7.61GPTFKK6 pKa = 10.87PNVSKK11 pKa = 10.88RR12 pKa = 11.84KK13 pKa = 8.93KK14 pKa = 8.63NHH16 pKa = 4.89GFRR19 pKa = 11.84KK20 pKa = 10.06RR21 pKa = 11.84MASKK25 pKa = 10.64NGRR28 pKa = 11.84KK29 pKa = 9.12VLAARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.23GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2016309 |
27 |
3561 |
329.9 |
36.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.744 ± 0.042 | 0.76 ± 0.01 |
5.421 ± 0.026 | 6.359 ± 0.039 |
3.902 ± 0.023 | 7.409 ± 0.029 |
2.072 ± 0.017 | 6.51 ± 0.032 |
4.732 ± 0.029 | 9.786 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.762 ± 0.02 | 3.876 ± 0.026 |
3.963 ± 0.021 | 3.93 ± 0.022 |
5.261 ± 0.036 | 6.66 ± 0.033 |
5.865 ± 0.048 | 7.128 ± 0.029 |
1.321 ± 0.014 | 3.539 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
