
Acetobacter nitrogenifigens DSM 23921 = LMG 23498
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Acetobacter; Acetobacter nitrogenifigens
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
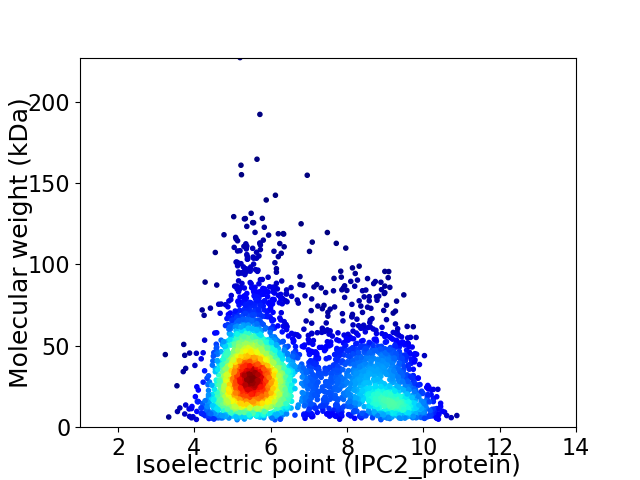
Virtual 2D-PAGE plot for 3660 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A511XDQ0|A0A511XDQ0_9PROT Cysteine synthase OS=Acetobacter nitrogenifigens DSM 23921 = LMG 23498 OX=1120919 GN=cysK1 PE=4 SV=1
MM1 pKa = 7.7SIFNSLTTAVAGINAQSTAFTNLSNNIANSQTTGYY36 pKa = 9.63KK37 pKa = 10.43ASTTSFQDD45 pKa = 3.73FVSSSLRR52 pKa = 11.84SSSSDD57 pKa = 3.06AVSDD61 pKa = 3.78GVGAVTHH68 pKa = 7.0TSTQQQGTVSASTNSLAVAISGNGFFDD95 pKa = 3.44VSKK98 pKa = 10.74EE99 pKa = 4.07SGQATSGTEE108 pKa = 3.69RR109 pKa = 11.84FSDD112 pKa = 3.35QQYY115 pKa = 7.59YY116 pKa = 8.48TRR118 pKa = 11.84NGDD121 pKa = 3.34FYY123 pKa = 11.12EE124 pKa = 4.65DD125 pKa = 3.41KK126 pKa = 11.01NGYY129 pKa = 8.78LVNTSGYY136 pKa = 10.45YY137 pKa = 10.1LDD139 pKa = 5.91GYY141 pKa = 9.95QADD144 pKa = 4.09ATTGTLGTSLTQINVSNVVFRR165 pKa = 11.84PTEE168 pKa = 4.13TTKK171 pKa = 10.36LTMTGDD177 pKa = 3.58VGSTTLPSSGAATTTTSSTIYY198 pKa = 10.18DD199 pKa = 3.84SQSASHH205 pKa = 6.27TLDD208 pKa = 3.38VTWTQDD214 pKa = 2.95SANPLKK220 pKa = 9.33WTVSASDD227 pKa = 4.34PSDD230 pKa = 3.28SSAISATPYY239 pKa = 9.99TVTFDD244 pKa = 3.31SSGAIASVTDD254 pKa = 3.43SSGDD258 pKa = 3.52AVGSSLSGATASIPITANYY277 pKa = 10.0NGTSQSIALNIGTIGSSSATTLSTTSSTSTTPTLTSDD314 pKa = 3.32SVTSGTYY321 pKa = 9.8EE322 pKa = 4.07GVEE325 pKa = 4.14MEE327 pKa = 4.28SDD329 pKa = 3.49GSVMAEE335 pKa = 4.11FSNGDD340 pKa = 3.36TQLVGKK346 pKa = 9.98IALSSFANADD356 pKa = 3.32GLSAVDD362 pKa = 3.56GQAYY366 pKa = 10.44VGTAQSGSATTGVVGANGTGTLEE389 pKa = 4.17TSSVEE394 pKa = 4.4SSTTDD399 pKa = 3.23LTSDD403 pKa = 3.71LSGLIVAQEE412 pKa = 4.2AYY414 pKa = 8.18TANTKK419 pKa = 9.92VVTTANQLLQSTIAMIQQ436 pKa = 2.9
MM1 pKa = 7.7SIFNSLTTAVAGINAQSTAFTNLSNNIANSQTTGYY36 pKa = 9.63KK37 pKa = 10.43ASTTSFQDD45 pKa = 3.73FVSSSLRR52 pKa = 11.84SSSSDD57 pKa = 3.06AVSDD61 pKa = 3.78GVGAVTHH68 pKa = 7.0TSTQQQGTVSASTNSLAVAISGNGFFDD95 pKa = 3.44VSKK98 pKa = 10.74EE99 pKa = 4.07SGQATSGTEE108 pKa = 3.69RR109 pKa = 11.84FSDD112 pKa = 3.35QQYY115 pKa = 7.59YY116 pKa = 8.48TRR118 pKa = 11.84NGDD121 pKa = 3.34FYY123 pKa = 11.12EE124 pKa = 4.65DD125 pKa = 3.41KK126 pKa = 11.01NGYY129 pKa = 8.78LVNTSGYY136 pKa = 10.45YY137 pKa = 10.1LDD139 pKa = 5.91GYY141 pKa = 9.95QADD144 pKa = 4.09ATTGTLGTSLTQINVSNVVFRR165 pKa = 11.84PTEE168 pKa = 4.13TTKK171 pKa = 10.36LTMTGDD177 pKa = 3.58VGSTTLPSSGAATTTTSSTIYY198 pKa = 10.18DD199 pKa = 3.84SQSASHH205 pKa = 6.27TLDD208 pKa = 3.38VTWTQDD214 pKa = 2.95SANPLKK220 pKa = 9.33WTVSASDD227 pKa = 4.34PSDD230 pKa = 3.28SSAISATPYY239 pKa = 9.99TVTFDD244 pKa = 3.31SSGAIASVTDD254 pKa = 3.43SSGDD258 pKa = 3.52AVGSSLSGATASIPITANYY277 pKa = 10.0NGTSQSIALNIGTIGSSSATTLSTTSSTSTTPTLTSDD314 pKa = 3.32SVTSGTYY321 pKa = 9.8EE322 pKa = 4.07GVEE325 pKa = 4.14MEE327 pKa = 4.28SDD329 pKa = 3.49GSVMAEE335 pKa = 4.11FSNGDD340 pKa = 3.36TQLVGKK346 pKa = 9.98IALSSFANADD356 pKa = 3.32GLSAVDD362 pKa = 3.56GQAYY366 pKa = 10.44VGTAQSGSATTGVVGANGTGTLEE389 pKa = 4.17TSSVEE394 pKa = 4.4SSTTDD399 pKa = 3.23LTSDD403 pKa = 3.71LSGLIVAQEE412 pKa = 4.2AYY414 pKa = 8.18TANTKK419 pKa = 9.92VVTTANQLLQSTIAMIQQ436 pKa = 2.9
Molecular weight: 44.15 kDa
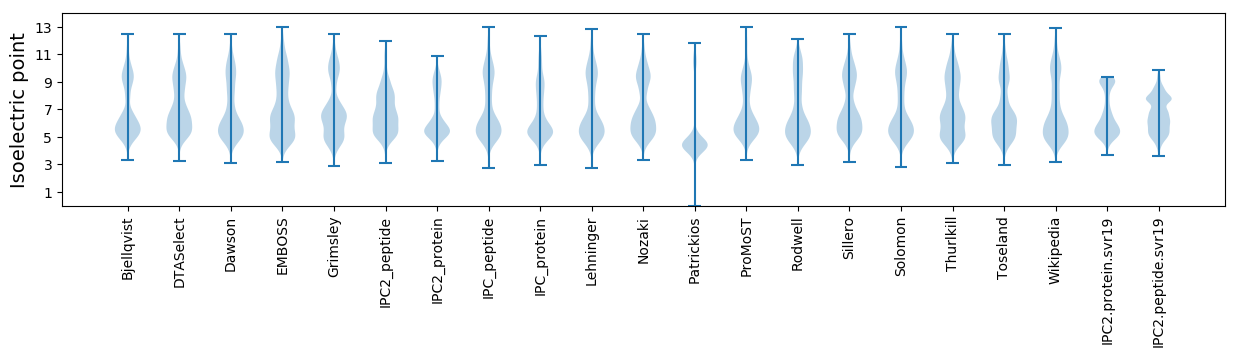
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A511X773|A0A511X773_9PROT Flavin prenyltransferase UbiX OS=Acetobacter nitrogenifigens DSM 23921 = LMG 23498 OX=1120919 GN=ubiX PE=3 SV=1
MM1 pKa = 7.26TLAPAQPSSVMPGRR15 pKa = 11.84DD16 pKa = 3.01WRR18 pKa = 11.84DD19 pKa = 3.01LRR21 pKa = 11.84PRR23 pKa = 11.84LLSALALIIVAVLAISLGGYY43 pKa = 10.34AYY45 pKa = 10.52GLLIIATMAGMAAEE59 pKa = 4.79GAALFALSPRR69 pKa = 11.84SWRR72 pKa = 11.84GALFVAWSFCAGLSAFCGRR91 pKa = 11.84WEE93 pKa = 4.05ATPAFAMTAFVFGPPLWLMNAVIAAGGLSLMWLRR127 pKa = 11.84LGADD131 pKa = 3.46CGVWAVVFVIAVVVSSDD148 pKa = 2.85SAAYY152 pKa = 8.25VTGRR156 pKa = 11.84MIGGPKK162 pKa = 9.63LAPRR166 pKa = 11.84ISPGKK171 pKa = 7.59TRR173 pKa = 11.84SGAVGGLLGAVMAGLVVATISGAGFAARR201 pKa = 11.84GAGWALLLGIAAQAGDD217 pKa = 3.87LAEE220 pKa = 4.42SAVKK224 pKa = 10.33RR225 pKa = 11.84ARR227 pKa = 11.84GVKK230 pKa = 10.19DD231 pKa = 3.08SGRR234 pKa = 11.84LLPGHH239 pKa = 6.75GGLLDD244 pKa = 4.13RR245 pKa = 11.84FDD247 pKa = 5.04ALLAAAPLAALLSLLAKK264 pKa = 10.08PGLPFWSAGFDD275 pKa = 3.52DD276 pKa = 4.95LARR279 pKa = 11.84GLGRR283 pKa = 11.84FLGGG287 pKa = 3.36
MM1 pKa = 7.26TLAPAQPSSVMPGRR15 pKa = 11.84DD16 pKa = 3.01WRR18 pKa = 11.84DD19 pKa = 3.01LRR21 pKa = 11.84PRR23 pKa = 11.84LLSALALIIVAVLAISLGGYY43 pKa = 10.34AYY45 pKa = 10.52GLLIIATMAGMAAEE59 pKa = 4.79GAALFALSPRR69 pKa = 11.84SWRR72 pKa = 11.84GALFVAWSFCAGLSAFCGRR91 pKa = 11.84WEE93 pKa = 4.05ATPAFAMTAFVFGPPLWLMNAVIAAGGLSLMWLRR127 pKa = 11.84LGADD131 pKa = 3.46CGVWAVVFVIAVVVSSDD148 pKa = 2.85SAAYY152 pKa = 8.25VTGRR156 pKa = 11.84MIGGPKK162 pKa = 9.63LAPRR166 pKa = 11.84ISPGKK171 pKa = 7.59TRR173 pKa = 11.84SGAVGGLLGAVMAGLVVATISGAGFAARR201 pKa = 11.84GAGWALLLGIAAQAGDD217 pKa = 3.87LAEE220 pKa = 4.42SAVKK224 pKa = 10.33RR225 pKa = 11.84ARR227 pKa = 11.84GVKK230 pKa = 10.19DD231 pKa = 3.08SGRR234 pKa = 11.84LLPGHH239 pKa = 6.75GGLLDD244 pKa = 4.13RR245 pKa = 11.84FDD247 pKa = 5.04ALLAAAPLAALLSLLAKK264 pKa = 10.08PGLPFWSAGFDD275 pKa = 3.52DD276 pKa = 4.95LARR279 pKa = 11.84GLGRR283 pKa = 11.84FLGGG287 pKa = 3.36
Molecular weight: 29.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1164225 |
41 |
2112 |
318.1 |
34.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.561 ± 0.051 | 0.919 ± 0.012 |
5.661 ± 0.03 | 5.22 ± 0.04 |
3.556 ± 0.026 | 8.665 ± 0.036 |
2.168 ± 0.019 | 4.777 ± 0.027 |
2.716 ± 0.031 | 10.048 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.386 ± 0.017 | 2.632 ± 0.028 |
5.423 ± 0.032 | 3.075 ± 0.025 |
7.371 ± 0.044 | 6.119 ± 0.035 |
5.629 ± 0.031 | 7.452 ± 0.031 |
1.396 ± 0.016 | 2.226 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
