
Winogradskyella sp. PG-2
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Winogradskyella; unclassified Winogradskyella
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
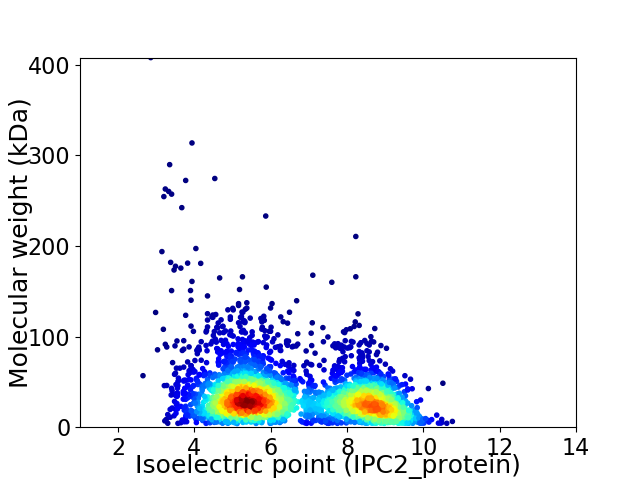
Virtual 2D-PAGE plot for 3520 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024FL28|A0A024FL28_9FLAO Uncharacterized protein OS=Winogradskyella sp. PG-2 OX=754409 GN=WPG_3344 PE=4 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84KK3 pKa = 8.93ILLFNVLCFVINNVFGQISLDD24 pKa = 3.62DD25 pKa = 3.66RR26 pKa = 11.84VNIYY30 pKa = 10.94DD31 pKa = 3.98FDD33 pKa = 4.26SLSVNIIYY41 pKa = 10.42DD42 pKa = 3.75DD43 pKa = 3.89FDD45 pKa = 5.24NDD47 pKa = 3.42GDD49 pKa = 4.3LDD51 pKa = 3.61IMKK54 pKa = 10.48FSGTDD59 pKa = 3.05SFNVLLQKK67 pKa = 10.86NNNGDD72 pKa = 3.62FNLNQPILVSSGVRR86 pKa = 11.84PIVSLDD92 pKa = 3.31LDD94 pKa = 3.62NDD96 pKa = 3.85GFLDD100 pKa = 3.9LLTYY104 pKa = 10.75HH105 pKa = 7.0SFTTIGVLYY114 pKa = 9.53NQQDD118 pKa = 3.49NTFSEE123 pKa = 4.88EE124 pKa = 4.01VTVQTFNGSYY134 pKa = 9.94DD135 pKa = 3.28IHH137 pKa = 6.91PMKK140 pKa = 10.46FDD142 pKa = 3.84YY143 pKa = 11.5NNDD146 pKa = 2.78GFMDD150 pKa = 5.67LIAIDD155 pKa = 4.62DD156 pKa = 4.12SEE158 pKa = 4.29DD159 pKa = 3.75AYY161 pKa = 11.88VLINNQMGGLEE172 pKa = 4.17PPQFLVSVGTFDD184 pKa = 5.01FLYY187 pKa = 11.05DD188 pKa = 5.32LDD190 pKa = 5.96DD191 pKa = 4.69FDD193 pKa = 6.32NDD195 pKa = 3.63GDD197 pKa = 3.86FDD199 pKa = 5.09FYY201 pKa = 11.05IRR203 pKa = 11.84DD204 pKa = 3.6GDD206 pKa = 3.93KK207 pKa = 11.24LIININGSGVFDD219 pKa = 3.83QPGSQQTQSSLRR231 pKa = 11.84SFEE234 pKa = 4.15ILDD237 pKa = 3.52IDD239 pKa = 3.92GNGYY243 pKa = 10.48KK244 pKa = 10.34DD245 pKa = 3.35VLYY248 pKa = 10.08WKK250 pKa = 10.51NDD252 pKa = 4.3AIWAKK257 pKa = 10.52YY258 pKa = 9.27YY259 pKa = 10.78GYY261 pKa = 10.87NATTTRR267 pKa = 11.84FIVINDD273 pKa = 3.29VMVVNNIPKK282 pKa = 10.04FSNYY286 pKa = 8.08VNARR290 pKa = 11.84SIHH293 pKa = 7.59IEE295 pKa = 3.95DD296 pKa = 3.95QDD298 pKa = 3.89NGSHH302 pKa = 6.87DD303 pKa = 4.12VYY305 pKa = 10.69VTLATIEE312 pKa = 4.37NQLDD316 pKa = 3.37MHH318 pKa = 6.92KK319 pKa = 10.88FNIQNNVFSNAQIVLSDD336 pKa = 3.29FQVNVFGVDD345 pKa = 3.03EE346 pKa = 5.26FSFLDD351 pKa = 4.83LNNDD355 pKa = 3.63DD356 pKa = 5.23NLDD359 pKa = 3.65FSFVSNFNQNKK370 pKa = 10.02FIFINNNINDD380 pKa = 4.3TPDD383 pKa = 2.96KK384 pKa = 10.2TICIQQAVKK393 pKa = 10.58PNDD396 pKa = 3.81FYY398 pKa = 11.79VIDD401 pKa = 3.89MNGDD405 pKa = 3.61GEE407 pKa = 4.53NDD409 pKa = 3.12ICVGTQNGLGYY420 pKa = 10.07FEE422 pKa = 4.38KK423 pKa = 10.04TASNEE428 pKa = 3.79LSGLRR433 pKa = 11.84SLIGVSTNPNASAYY447 pKa = 6.41THH449 pKa = 7.04INITDD454 pKa = 3.51INNDD458 pKa = 3.16GLGDD462 pKa = 3.88VIDD465 pKa = 3.77YY466 pKa = 10.08TGFDD470 pKa = 3.49NEE472 pKa = 4.43AKK474 pKa = 10.19IFKK477 pKa = 10.5NLGDD481 pKa = 4.17DD482 pKa = 3.24NFEE485 pKa = 4.28FVQSVSMTGIFNGTHH500 pKa = 6.82LSFADD505 pKa = 3.64IDD507 pKa = 4.12GDD509 pKa = 3.98DD510 pKa = 4.37FKK512 pKa = 11.78DD513 pKa = 4.51LVIFDD518 pKa = 3.88QPNFYY523 pKa = 8.78WAKK526 pKa = 10.11NNNGLSFQSLQQLVVNNVDD545 pKa = 3.31NDD547 pKa = 3.98DD548 pKa = 4.65PLSIMYY554 pKa = 10.42EE555 pKa = 4.02DD556 pKa = 5.25FNDD559 pKa = 4.15DD560 pKa = 4.02DD561 pKa = 5.44EE562 pKa = 6.6IDD564 pKa = 3.8ILVLRR569 pKa = 11.84YY570 pKa = 9.25FYY572 pKa = 10.97EE573 pKa = 4.2SGQFHH578 pKa = 6.3TEE580 pKa = 3.77VNLLEE585 pKa = 4.35NDD587 pKa = 3.26NGEE590 pKa = 4.07FTGNIIADD598 pKa = 3.57FNGNYY603 pKa = 9.25GRR605 pKa = 11.84SHH607 pKa = 6.99LKK609 pKa = 9.91IDD611 pKa = 5.96DD612 pKa = 3.95FDD614 pKa = 3.91QDD616 pKa = 2.99NDD618 pKa = 3.35LDD620 pKa = 4.31FFVHH624 pKa = 5.76STRR627 pKa = 11.84LDD629 pKa = 3.42QPLLFFKK636 pKa = 11.01NDD638 pKa = 2.95GSNNFISTTIDD649 pKa = 3.42NIDD652 pKa = 3.33IEE654 pKa = 5.54DD655 pKa = 3.53IEE657 pKa = 5.58FYY659 pKa = 11.08DD660 pKa = 5.13GDD662 pKa = 3.74GDD664 pKa = 4.38GYY666 pKa = 11.45NEE668 pKa = 3.8IYY670 pKa = 10.02AWNYY674 pKa = 10.01EE675 pKa = 4.05SFANHH680 pKa = 5.94IFYY683 pKa = 10.85YY684 pKa = 10.0DD685 pKa = 3.26TTDD688 pKa = 3.51YY689 pKa = 11.54INFTKK694 pKa = 10.3IPIDD698 pKa = 3.88SYY700 pKa = 11.17SASYY704 pKa = 11.32DD705 pKa = 3.41NLDD708 pKa = 3.78DD709 pKa = 3.78YY710 pKa = 11.83TRR712 pKa = 11.84GDD714 pKa = 3.74LFMYY718 pKa = 10.18DD719 pKa = 3.82YY720 pKa = 11.76NNDD723 pKa = 3.36GKK725 pKa = 10.76DD726 pKa = 3.41DD727 pKa = 4.31LFINNYY733 pKa = 10.29SSFQALISVYY743 pKa = 10.31EE744 pKa = 4.09NTSEE748 pKa = 4.11SLGVEE753 pKa = 4.07EE754 pKa = 5.24IDD756 pKa = 4.46NNSLSQMRR764 pKa = 11.84LYY766 pKa = 10.33PNPFVNSVKK775 pKa = 8.1WTKK778 pKa = 10.43RR779 pKa = 11.84GNEE782 pKa = 4.14TYY784 pKa = 10.85SLQLYY789 pKa = 9.12SQDD792 pKa = 2.79GKK794 pKa = 11.48LIFEE798 pKa = 4.63KK799 pKa = 10.82VISEE803 pKa = 4.11NNMDD807 pKa = 4.12FSSYY811 pKa = 11.24DD812 pKa = 3.12SGVYY816 pKa = 9.58FLSISEE822 pKa = 4.39VISGNKK828 pKa = 7.89KK829 pKa = 6.1TFKK832 pKa = 10.28IIKK835 pKa = 9.12NN836 pKa = 3.59
MM1 pKa = 7.97RR2 pKa = 11.84KK3 pKa = 8.93ILLFNVLCFVINNVFGQISLDD24 pKa = 3.62DD25 pKa = 3.66RR26 pKa = 11.84VNIYY30 pKa = 10.94DD31 pKa = 3.98FDD33 pKa = 4.26SLSVNIIYY41 pKa = 10.42DD42 pKa = 3.75DD43 pKa = 3.89FDD45 pKa = 5.24NDD47 pKa = 3.42GDD49 pKa = 4.3LDD51 pKa = 3.61IMKK54 pKa = 10.48FSGTDD59 pKa = 3.05SFNVLLQKK67 pKa = 10.86NNNGDD72 pKa = 3.62FNLNQPILVSSGVRR86 pKa = 11.84PIVSLDD92 pKa = 3.31LDD94 pKa = 3.62NDD96 pKa = 3.85GFLDD100 pKa = 3.9LLTYY104 pKa = 10.75HH105 pKa = 7.0SFTTIGVLYY114 pKa = 9.53NQQDD118 pKa = 3.49NTFSEE123 pKa = 4.88EE124 pKa = 4.01VTVQTFNGSYY134 pKa = 9.94DD135 pKa = 3.28IHH137 pKa = 6.91PMKK140 pKa = 10.46FDD142 pKa = 3.84YY143 pKa = 11.5NNDD146 pKa = 2.78GFMDD150 pKa = 5.67LIAIDD155 pKa = 4.62DD156 pKa = 4.12SEE158 pKa = 4.29DD159 pKa = 3.75AYY161 pKa = 11.88VLINNQMGGLEE172 pKa = 4.17PPQFLVSVGTFDD184 pKa = 5.01FLYY187 pKa = 11.05DD188 pKa = 5.32LDD190 pKa = 5.96DD191 pKa = 4.69FDD193 pKa = 6.32NDD195 pKa = 3.63GDD197 pKa = 3.86FDD199 pKa = 5.09FYY201 pKa = 11.05IRR203 pKa = 11.84DD204 pKa = 3.6GDD206 pKa = 3.93KK207 pKa = 11.24LIININGSGVFDD219 pKa = 3.83QPGSQQTQSSLRR231 pKa = 11.84SFEE234 pKa = 4.15ILDD237 pKa = 3.52IDD239 pKa = 3.92GNGYY243 pKa = 10.48KK244 pKa = 10.34DD245 pKa = 3.35VLYY248 pKa = 10.08WKK250 pKa = 10.51NDD252 pKa = 4.3AIWAKK257 pKa = 10.52YY258 pKa = 9.27YY259 pKa = 10.78GYY261 pKa = 10.87NATTTRR267 pKa = 11.84FIVINDD273 pKa = 3.29VMVVNNIPKK282 pKa = 10.04FSNYY286 pKa = 8.08VNARR290 pKa = 11.84SIHH293 pKa = 7.59IEE295 pKa = 3.95DD296 pKa = 3.95QDD298 pKa = 3.89NGSHH302 pKa = 6.87DD303 pKa = 4.12VYY305 pKa = 10.69VTLATIEE312 pKa = 4.37NQLDD316 pKa = 3.37MHH318 pKa = 6.92KK319 pKa = 10.88FNIQNNVFSNAQIVLSDD336 pKa = 3.29FQVNVFGVDD345 pKa = 3.03EE346 pKa = 5.26FSFLDD351 pKa = 4.83LNNDD355 pKa = 3.63DD356 pKa = 5.23NLDD359 pKa = 3.65FSFVSNFNQNKK370 pKa = 10.02FIFINNNINDD380 pKa = 4.3TPDD383 pKa = 2.96KK384 pKa = 10.2TICIQQAVKK393 pKa = 10.58PNDD396 pKa = 3.81FYY398 pKa = 11.79VIDD401 pKa = 3.89MNGDD405 pKa = 3.61GEE407 pKa = 4.53NDD409 pKa = 3.12ICVGTQNGLGYY420 pKa = 10.07FEE422 pKa = 4.38KK423 pKa = 10.04TASNEE428 pKa = 3.79LSGLRR433 pKa = 11.84SLIGVSTNPNASAYY447 pKa = 6.41THH449 pKa = 7.04INITDD454 pKa = 3.51INNDD458 pKa = 3.16GLGDD462 pKa = 3.88VIDD465 pKa = 3.77YY466 pKa = 10.08TGFDD470 pKa = 3.49NEE472 pKa = 4.43AKK474 pKa = 10.19IFKK477 pKa = 10.5NLGDD481 pKa = 4.17DD482 pKa = 3.24NFEE485 pKa = 4.28FVQSVSMTGIFNGTHH500 pKa = 6.82LSFADD505 pKa = 3.64IDD507 pKa = 4.12GDD509 pKa = 3.98DD510 pKa = 4.37FKK512 pKa = 11.78DD513 pKa = 4.51LVIFDD518 pKa = 3.88QPNFYY523 pKa = 8.78WAKK526 pKa = 10.11NNNGLSFQSLQQLVVNNVDD545 pKa = 3.31NDD547 pKa = 3.98DD548 pKa = 4.65PLSIMYY554 pKa = 10.42EE555 pKa = 4.02DD556 pKa = 5.25FNDD559 pKa = 4.15DD560 pKa = 4.02DD561 pKa = 5.44EE562 pKa = 6.6IDD564 pKa = 3.8ILVLRR569 pKa = 11.84YY570 pKa = 9.25FYY572 pKa = 10.97EE573 pKa = 4.2SGQFHH578 pKa = 6.3TEE580 pKa = 3.77VNLLEE585 pKa = 4.35NDD587 pKa = 3.26NGEE590 pKa = 4.07FTGNIIADD598 pKa = 3.57FNGNYY603 pKa = 9.25GRR605 pKa = 11.84SHH607 pKa = 6.99LKK609 pKa = 9.91IDD611 pKa = 5.96DD612 pKa = 3.95FDD614 pKa = 3.91QDD616 pKa = 2.99NDD618 pKa = 3.35LDD620 pKa = 4.31FFVHH624 pKa = 5.76STRR627 pKa = 11.84LDD629 pKa = 3.42QPLLFFKK636 pKa = 11.01NDD638 pKa = 2.95GSNNFISTTIDD649 pKa = 3.42NIDD652 pKa = 3.33IEE654 pKa = 5.54DD655 pKa = 3.53IEE657 pKa = 5.58FYY659 pKa = 11.08DD660 pKa = 5.13GDD662 pKa = 3.74GDD664 pKa = 4.38GYY666 pKa = 11.45NEE668 pKa = 3.8IYY670 pKa = 10.02AWNYY674 pKa = 10.01EE675 pKa = 4.05SFANHH680 pKa = 5.94IFYY683 pKa = 10.85YY684 pKa = 10.0DD685 pKa = 3.26TTDD688 pKa = 3.51YY689 pKa = 11.54INFTKK694 pKa = 10.3IPIDD698 pKa = 3.88SYY700 pKa = 11.17SASYY704 pKa = 11.32DD705 pKa = 3.41NLDD708 pKa = 3.78DD709 pKa = 3.78YY710 pKa = 11.83TRR712 pKa = 11.84GDD714 pKa = 3.74LFMYY718 pKa = 10.18DD719 pKa = 3.82YY720 pKa = 11.76NNDD723 pKa = 3.36GKK725 pKa = 10.76DD726 pKa = 3.41DD727 pKa = 4.31LFINNYY733 pKa = 10.29SSFQALISVYY743 pKa = 10.31EE744 pKa = 4.09NTSEE748 pKa = 4.11SLGVEE753 pKa = 4.07EE754 pKa = 5.24IDD756 pKa = 4.46NNSLSQMRR764 pKa = 11.84LYY766 pKa = 10.33PNPFVNSVKK775 pKa = 8.1WTKK778 pKa = 10.43RR779 pKa = 11.84GNEE782 pKa = 4.14TYY784 pKa = 10.85SLQLYY789 pKa = 9.12SQDD792 pKa = 2.79GKK794 pKa = 11.48LIFEE798 pKa = 4.63KK799 pKa = 10.82VISEE803 pKa = 4.11NNMDD807 pKa = 4.12FSSYY811 pKa = 11.24DD812 pKa = 3.12SGVYY816 pKa = 9.58FLSISEE822 pKa = 4.39VISGNKK828 pKa = 7.89KK829 pKa = 6.1TFKK832 pKa = 10.28IIKK835 pKa = 9.12NN836 pKa = 3.59
Molecular weight: 95.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024FKC4|A0A024FKC4_9FLAO Diacylglycerol kinase OS=Winogradskyella sp. PG-2 OX=754409 GN=WPG_2743 PE=3 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.04TRR50 pKa = 11.84HH51 pKa = 5.93KK52 pKa = 10.8RR53 pKa = 3.27
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.05RR22 pKa = 11.84MASVNGRR29 pKa = 11.84KK30 pKa = 9.21VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.04TRR50 pKa = 11.84HH51 pKa = 5.93KK52 pKa = 10.8RR53 pKa = 3.27
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1145739 |
37 |
3972 |
325.5 |
36.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.043 ± 0.037 | 0.738 ± 0.015 |
5.998 ± 0.048 | 6.407 ± 0.04 |
5.259 ± 0.037 | 6.286 ± 0.046 |
1.672 ± 0.021 | 8.29 ± 0.039 |
7.696 ± 0.081 | 9.201 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.094 ± 0.025 | 6.677 ± 0.046 |
3.189 ± 0.028 | 3.158 ± 0.022 |
3.308 ± 0.036 | 6.794 ± 0.038 |
5.913 ± 0.062 | 6.042 ± 0.032 |
1.042 ± 0.014 | 4.196 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
