
Cohnella sp. CC-MHH1044
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Cohnella
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
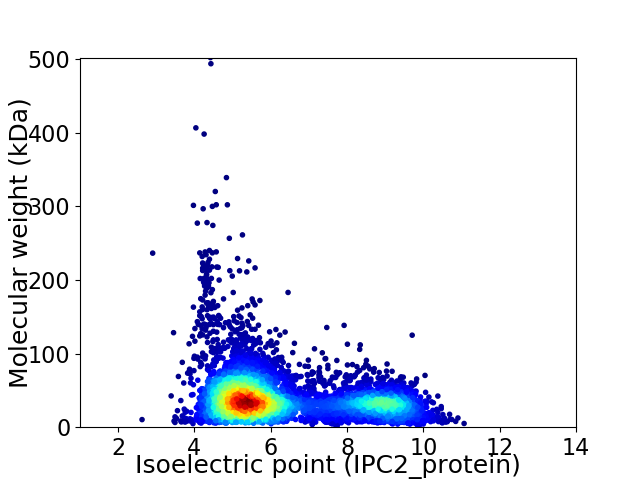
Virtual 2D-PAGE plot for 6462 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
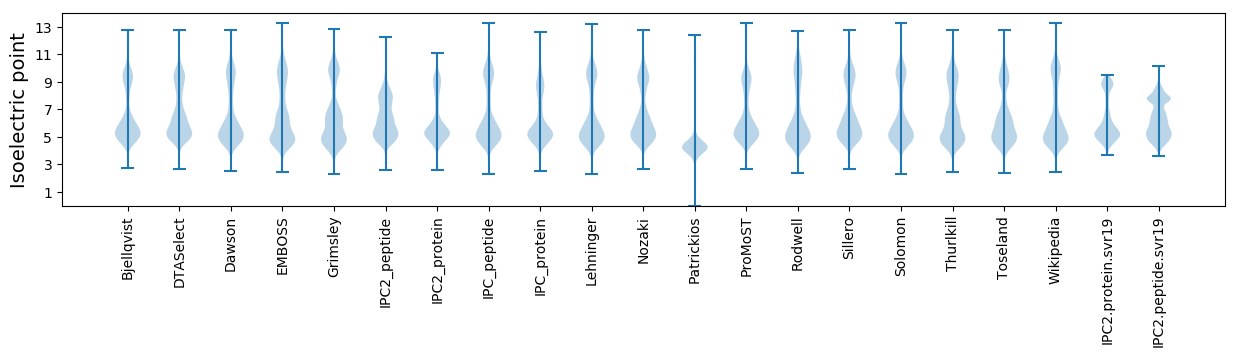
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S4BN37|A0A4S4BN37_9BACL LLM class flavin-dependent oxidoreductase OS=Cohnella sp. CC-MHH1044 OX=2565925 GN=E6C55_22090 PE=4 SV=1
MM1 pKa = 6.93VAHH4 pKa = 7.4LIGGYY9 pKa = 9.78SVTKK13 pKa = 10.18GKK15 pKa = 9.61WIGFGLLGLLALFVIWLFIPSEE37 pKa = 4.25DD38 pKa = 3.78TDD40 pKa = 3.52NTNVKK45 pKa = 10.27RR46 pKa = 11.84EE47 pKa = 3.88AAEE50 pKa = 3.75NDD52 pKa = 3.1GDD54 pKa = 4.22YY55 pKa = 11.1TILVYY60 pKa = 10.63MNGSDD65 pKa = 4.91LEE67 pKa = 4.23SEE69 pKa = 4.27YY70 pKa = 11.29DD71 pKa = 3.73EE72 pKa = 5.9EE73 pKa = 5.33SDD75 pKa = 3.87SYY77 pKa = 11.69YY78 pKa = 11.19GAASSDD84 pKa = 3.98LEE86 pKa = 4.14EE87 pKa = 4.82MISGLSGDD95 pKa = 3.69RR96 pKa = 11.84VRR98 pKa = 11.84VLVEE102 pKa = 3.84TGGTLAWADD111 pKa = 3.52EE112 pKa = 4.8RR113 pKa = 11.84IDD115 pKa = 4.6AEE117 pKa = 4.41QNQRR121 pKa = 11.84WLIEE125 pKa = 4.71DD126 pKa = 3.7GQFTHH131 pKa = 6.98AADD134 pKa = 3.3VGLRR138 pKa = 11.84NIGEE142 pKa = 4.64ADD144 pKa = 3.41TLADD148 pKa = 4.35FVAWGVEE155 pKa = 3.94QYY157 pKa = 10.64PADD160 pKa = 3.92KK161 pKa = 10.57YY162 pKa = 11.78ALIFWNHH169 pKa = 5.72GGGSVLGFGADD180 pKa = 3.04EE181 pKa = 4.49RR182 pKa = 11.84FDD184 pKa = 4.2YY185 pKa = 11.15DD186 pKa = 4.92SLTLDD191 pKa = 3.67EE192 pKa = 5.12LAAGLKK198 pKa = 7.08TAQMSTDD205 pKa = 2.94VSFEE209 pKa = 4.14VIGFDD214 pKa = 3.36ACLMANVEE222 pKa = 4.49TASLLSPYY230 pKa = 10.17GRR232 pKa = 11.84YY233 pKa = 9.18LVASEE238 pKa = 4.26EE239 pKa = 4.33LEE241 pKa = 4.55PGHH244 pKa = 6.16GWDD247 pKa = 3.45YY248 pKa = 10.36TAALSRR254 pKa = 11.84LSDD257 pKa = 3.6EE258 pKa = 4.74PDD260 pKa = 3.24ADD262 pKa = 3.96GATLGRR268 pKa = 11.84WIADD272 pKa = 4.06GYY274 pKa = 9.46RR275 pKa = 11.84QHH277 pKa = 8.41AEE279 pKa = 4.61DD280 pKa = 4.82NDD282 pKa = 3.63QEE284 pKa = 4.56KK285 pKa = 11.06NITLSVVDD293 pKa = 4.07MSRR296 pKa = 11.84MEE298 pKa = 4.22RR299 pKa = 11.84VVTALEE305 pKa = 4.22AFAQEE310 pKa = 4.09VDD312 pKa = 3.37SGISVDD318 pKa = 3.3RR319 pKa = 11.84SGFFSLANGRR329 pKa = 11.84SRR331 pKa = 11.84ADD333 pKa = 3.43EE334 pKa = 4.43YY335 pKa = 11.5GSSSTPEE342 pKa = 3.95EE343 pKa = 4.05ASDD346 pKa = 4.04MVDD349 pKa = 3.28LLAFVRR355 pKa = 11.84NVAEE359 pKa = 4.86RR360 pKa = 11.84YY361 pKa = 8.45PDD363 pKa = 3.37TSKK366 pKa = 10.96EE367 pKa = 4.04LEE369 pKa = 4.18NALQDD374 pKa = 3.41AVVYY378 pKa = 10.28NIVSVGHH385 pKa = 7.13PDD387 pKa = 3.22ASGLSIYY394 pKa = 10.24FPHH397 pKa = 6.99RR398 pKa = 11.84NKK400 pKa = 10.38INFDD404 pKa = 3.97DD405 pKa = 3.83NLQRR409 pKa = 11.84FAAIGFSDD417 pKa = 3.83KK418 pKa = 10.96YY419 pKa = 11.45VEE421 pKa = 3.95FLHH424 pKa = 6.65WYY426 pKa = 9.96VSGLLGNSEE435 pKa = 4.37TVEE438 pKa = 4.15VSTSGQLNFSYY449 pKa = 10.98NEE451 pKa = 4.11EE452 pKa = 3.89EE453 pKa = 5.15DD454 pKa = 4.01EE455 pKa = 4.24YY456 pKa = 11.59APYY459 pKa = 10.16EE460 pKa = 4.41VYY462 pKa = 10.83VDD464 pKa = 3.92PEE466 pKa = 3.99EE467 pKa = 5.1LEE469 pKa = 4.25QIEE472 pKa = 4.31QIYY475 pKa = 10.39AIVSMYY481 pKa = 10.64SDD483 pKa = 3.52EE484 pKa = 5.13DD485 pKa = 3.71AGEE488 pKa = 4.47DD489 pKa = 3.98SPLIYY494 pKa = 10.28LGYY497 pKa = 10.52DD498 pKa = 3.07HH499 pKa = 6.96YY500 pKa = 12.08VDD502 pKa = 4.12VDD504 pKa = 3.39WEE506 pKa = 4.13EE507 pKa = 4.46GIIRR511 pKa = 11.84DD512 pKa = 4.23DD513 pKa = 4.35FTGEE517 pKa = 3.16WLMWDD522 pKa = 3.83GNFVTLDD529 pKa = 3.84LVSQGDD535 pKa = 3.75GFIRR539 pKa = 11.84YY540 pKa = 8.4AIPAKK545 pKa = 10.84LNGKK549 pKa = 9.45DD550 pKa = 3.26VDD552 pKa = 3.84ILVHH556 pKa = 6.23YY557 pKa = 10.14DD558 pKa = 3.29VEE560 pKa = 4.51EE561 pKa = 4.05EE562 pKa = 4.17TFEE565 pKa = 5.42VLGAWKK571 pKa = 10.48GLGSEE576 pKa = 4.41TGMPDD581 pKa = 3.51KK582 pKa = 11.05NIIPIRR588 pKa = 11.84EE589 pKa = 3.9GDD591 pKa = 3.66RR592 pKa = 11.84IVPQFYY598 pKa = 10.12YY599 pKa = 10.66YY600 pKa = 10.64YY601 pKa = 10.8DD602 pKa = 3.84SGEE605 pKa = 4.21DD606 pKa = 3.57GYY608 pKa = 11.11MDD610 pKa = 3.67GDD612 pKa = 3.89EE613 pKa = 4.61FVVGDD618 pKa = 4.52TIDD621 pKa = 4.26LEE623 pKa = 4.26YY624 pKa = 10.75DD625 pKa = 3.46YY626 pKa = 10.97LPNGSYY632 pKa = 10.85LYY634 pKa = 10.74GFSLVSYY641 pKa = 10.07SGEE644 pKa = 3.85EE645 pKa = 3.89TLTDD649 pKa = 4.68FIEE652 pKa = 5.29FEE654 pKa = 4.73LEE656 pKa = 3.94DD657 pKa = 3.54
MM1 pKa = 6.93VAHH4 pKa = 7.4LIGGYY9 pKa = 9.78SVTKK13 pKa = 10.18GKK15 pKa = 9.61WIGFGLLGLLALFVIWLFIPSEE37 pKa = 4.25DD38 pKa = 3.78TDD40 pKa = 3.52NTNVKK45 pKa = 10.27RR46 pKa = 11.84EE47 pKa = 3.88AAEE50 pKa = 3.75NDD52 pKa = 3.1GDD54 pKa = 4.22YY55 pKa = 11.1TILVYY60 pKa = 10.63MNGSDD65 pKa = 4.91LEE67 pKa = 4.23SEE69 pKa = 4.27YY70 pKa = 11.29DD71 pKa = 3.73EE72 pKa = 5.9EE73 pKa = 5.33SDD75 pKa = 3.87SYY77 pKa = 11.69YY78 pKa = 11.19GAASSDD84 pKa = 3.98LEE86 pKa = 4.14EE87 pKa = 4.82MISGLSGDD95 pKa = 3.69RR96 pKa = 11.84VRR98 pKa = 11.84VLVEE102 pKa = 3.84TGGTLAWADD111 pKa = 3.52EE112 pKa = 4.8RR113 pKa = 11.84IDD115 pKa = 4.6AEE117 pKa = 4.41QNQRR121 pKa = 11.84WLIEE125 pKa = 4.71DD126 pKa = 3.7GQFTHH131 pKa = 6.98AADD134 pKa = 3.3VGLRR138 pKa = 11.84NIGEE142 pKa = 4.64ADD144 pKa = 3.41TLADD148 pKa = 4.35FVAWGVEE155 pKa = 3.94QYY157 pKa = 10.64PADD160 pKa = 3.92KK161 pKa = 10.57YY162 pKa = 11.78ALIFWNHH169 pKa = 5.72GGGSVLGFGADD180 pKa = 3.04EE181 pKa = 4.49RR182 pKa = 11.84FDD184 pKa = 4.2YY185 pKa = 11.15DD186 pKa = 4.92SLTLDD191 pKa = 3.67EE192 pKa = 5.12LAAGLKK198 pKa = 7.08TAQMSTDD205 pKa = 2.94VSFEE209 pKa = 4.14VIGFDD214 pKa = 3.36ACLMANVEE222 pKa = 4.49TASLLSPYY230 pKa = 10.17GRR232 pKa = 11.84YY233 pKa = 9.18LVASEE238 pKa = 4.26EE239 pKa = 4.33LEE241 pKa = 4.55PGHH244 pKa = 6.16GWDD247 pKa = 3.45YY248 pKa = 10.36TAALSRR254 pKa = 11.84LSDD257 pKa = 3.6EE258 pKa = 4.74PDD260 pKa = 3.24ADD262 pKa = 3.96GATLGRR268 pKa = 11.84WIADD272 pKa = 4.06GYY274 pKa = 9.46RR275 pKa = 11.84QHH277 pKa = 8.41AEE279 pKa = 4.61DD280 pKa = 4.82NDD282 pKa = 3.63QEE284 pKa = 4.56KK285 pKa = 11.06NITLSVVDD293 pKa = 4.07MSRR296 pKa = 11.84MEE298 pKa = 4.22RR299 pKa = 11.84VVTALEE305 pKa = 4.22AFAQEE310 pKa = 4.09VDD312 pKa = 3.37SGISVDD318 pKa = 3.3RR319 pKa = 11.84SGFFSLANGRR329 pKa = 11.84SRR331 pKa = 11.84ADD333 pKa = 3.43EE334 pKa = 4.43YY335 pKa = 11.5GSSSTPEE342 pKa = 3.95EE343 pKa = 4.05ASDD346 pKa = 4.04MVDD349 pKa = 3.28LLAFVRR355 pKa = 11.84NVAEE359 pKa = 4.86RR360 pKa = 11.84YY361 pKa = 8.45PDD363 pKa = 3.37TSKK366 pKa = 10.96EE367 pKa = 4.04LEE369 pKa = 4.18NALQDD374 pKa = 3.41AVVYY378 pKa = 10.28NIVSVGHH385 pKa = 7.13PDD387 pKa = 3.22ASGLSIYY394 pKa = 10.24FPHH397 pKa = 6.99RR398 pKa = 11.84NKK400 pKa = 10.38INFDD404 pKa = 3.97DD405 pKa = 3.83NLQRR409 pKa = 11.84FAAIGFSDD417 pKa = 3.83KK418 pKa = 10.96YY419 pKa = 11.45VEE421 pKa = 3.95FLHH424 pKa = 6.65WYY426 pKa = 9.96VSGLLGNSEE435 pKa = 4.37TVEE438 pKa = 4.15VSTSGQLNFSYY449 pKa = 10.98NEE451 pKa = 4.11EE452 pKa = 3.89EE453 pKa = 5.15DD454 pKa = 4.01EE455 pKa = 4.24YY456 pKa = 11.59APYY459 pKa = 10.16EE460 pKa = 4.41VYY462 pKa = 10.83VDD464 pKa = 3.92PEE466 pKa = 3.99EE467 pKa = 5.1LEE469 pKa = 4.25QIEE472 pKa = 4.31QIYY475 pKa = 10.39AIVSMYY481 pKa = 10.64SDD483 pKa = 3.52EE484 pKa = 5.13DD485 pKa = 3.71AGEE488 pKa = 4.47DD489 pKa = 3.98SPLIYY494 pKa = 10.28LGYY497 pKa = 10.52DD498 pKa = 3.07HH499 pKa = 6.96YY500 pKa = 12.08VDD502 pKa = 4.12VDD504 pKa = 3.39WEE506 pKa = 4.13EE507 pKa = 4.46GIIRR511 pKa = 11.84DD512 pKa = 4.23DD513 pKa = 4.35FTGEE517 pKa = 3.16WLMWDD522 pKa = 3.83GNFVTLDD529 pKa = 3.84LVSQGDD535 pKa = 3.75GFIRR539 pKa = 11.84YY540 pKa = 8.4AIPAKK545 pKa = 10.84LNGKK549 pKa = 9.45DD550 pKa = 3.26VDD552 pKa = 3.84ILVHH556 pKa = 6.23YY557 pKa = 10.14DD558 pKa = 3.29VEE560 pKa = 4.51EE561 pKa = 4.05EE562 pKa = 4.17TFEE565 pKa = 5.42VLGAWKK571 pKa = 10.48GLGSEE576 pKa = 4.41TGMPDD581 pKa = 3.51KK582 pKa = 11.05NIIPIRR588 pKa = 11.84EE589 pKa = 3.9GDD591 pKa = 3.66RR592 pKa = 11.84IVPQFYY598 pKa = 10.12YY599 pKa = 10.66YY600 pKa = 10.64YY601 pKa = 10.8DD602 pKa = 3.84SGEE605 pKa = 4.21DD606 pKa = 3.57GYY608 pKa = 11.11MDD610 pKa = 3.67GDD612 pKa = 3.89EE613 pKa = 4.61FVVGDD618 pKa = 4.52TIDD621 pKa = 4.26LEE623 pKa = 4.26YY624 pKa = 10.75DD625 pKa = 3.46YY626 pKa = 10.97LPNGSYY632 pKa = 10.85LYY634 pKa = 10.74GFSLVSYY641 pKa = 10.07SGEE644 pKa = 3.85EE645 pKa = 3.89TLTDD649 pKa = 4.68FIEE652 pKa = 5.29FEE654 pKa = 4.73LEE656 pKa = 3.94DD657 pKa = 3.54
Molecular weight: 73.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S4BIX2|A0A4S4BIX2_9BACL Carb-bd_dom_fam9 domain-containing protein OS=Cohnella sp. CC-MHH1044 OX=2565925 GN=E6C55_24620 PE=4 SV=1
MM1 pKa = 7.61GPTFKK6 pKa = 10.87PNVSKK11 pKa = 10.8RR12 pKa = 11.84KK13 pKa = 8.57KK14 pKa = 8.17VHH16 pKa = 5.5GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSKK25 pKa = 10.58NGRR28 pKa = 11.84KK29 pKa = 8.51VLKK32 pKa = 10.34ARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.21GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
MM1 pKa = 7.61GPTFKK6 pKa = 10.87PNVSKK11 pKa = 10.8RR12 pKa = 11.84KK13 pKa = 8.57KK14 pKa = 8.17VHH16 pKa = 5.5GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSKK25 pKa = 10.58NGRR28 pKa = 11.84KK29 pKa = 8.51VLKK32 pKa = 10.34ARR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.21GRR39 pKa = 11.84KK40 pKa = 8.56VLSAA44 pKa = 4.05
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2442047 |
24 |
4735 |
377.9 |
41.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.687 ± 0.046 | 0.711 ± 0.009 |
5.4 ± 0.023 | 6.736 ± 0.037 |
3.879 ± 0.021 | 8.074 ± 0.037 |
1.795 ± 0.016 | 5.697 ± 0.026 |
4.269 ± 0.029 | 10.165 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.444 ± 0.014 | 3.514 ± 0.027 |
4.275 ± 0.019 | 3.341 ± 0.017 |
5.991 ± 0.039 | 6.645 ± 0.03 |
5.454 ± 0.037 | 6.954 ± 0.026 |
1.432 ± 0.013 | 3.539 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
