
Sulfuritalea hydrogenivorans sk43H
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Sterolibacteriaceae; Sulfuritalea; Sulfuritalea hydrogenivorans
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
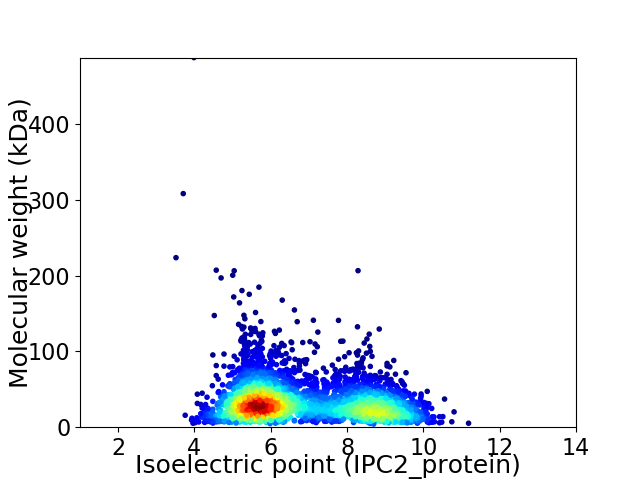
Virtual 2D-PAGE plot for 3578 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
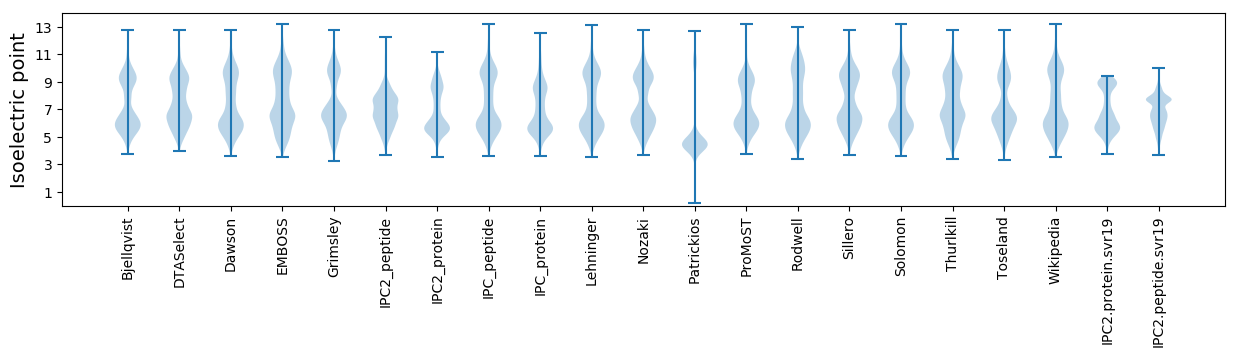
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0SGV9|W0SGV9_9PROT 3-oxoacyl-[acyl-carrier-protein] reductase OS=Sulfuritalea hydrogenivorans sk43H OX=1223802 GN=fabG PE=3 SV=1
MM1 pKa = 7.7GNFDD5 pKa = 3.99LHH7 pKa = 7.65GLINAIANNHH17 pKa = 5.08GVASPVKK24 pKa = 8.74SAKK27 pKa = 10.04RR28 pKa = 11.84KK29 pKa = 8.43MRR31 pKa = 11.84DD32 pKa = 3.02VSEE35 pKa = 4.82GDD37 pKa = 3.59GQATAQAEE45 pKa = 4.23GDD47 pKa = 3.79ATSATSDD54 pKa = 3.18ASMQLAQADD63 pKa = 3.76TGVATEE69 pKa = 4.24AAAGEE74 pKa = 4.54TTGGASGAPAAGEE87 pKa = 3.84AAAGSGAAAGGLGGLGLGGMALGGAALAGGGGGGGSAVAAAVNNIITGTVVAGPVLAGSGLKK149 pKa = 10.26VEE151 pKa = 5.26IYY153 pKa = 10.3AANGTTKK160 pKa = 10.58LGEE163 pKa = 4.29STLDD167 pKa = 3.2ASGKK171 pKa = 8.51FTVDD175 pKa = 2.53VGAYY179 pKa = 7.71TGIVIARR186 pKa = 11.84LVDD189 pKa = 3.69ANGGDD194 pKa = 4.09DD195 pKa = 4.33YY196 pKa = 11.62LDD198 pKa = 3.91EE199 pKa = 4.51ATGVPKK205 pKa = 10.61DD206 pKa = 3.7LNANLMATSVVTGGTFTLNINPLTTIAAQKK236 pKa = 10.66AGLAADD242 pKa = 4.57GSGGLADD249 pKa = 4.07AAAVANANAAVAAAFGLTDD268 pKa = 3.61LTGTSVVATNGGSYY282 pKa = 10.69DD283 pKa = 3.6AADD286 pKa = 3.51GMSAGEE292 pKa = 4.2KK293 pKa = 10.26YY294 pKa = 10.62GAILAALSGADD305 pKa = 3.73FASSGDD311 pKa = 3.62SQAIIDD317 pKa = 4.25NLVAAIDD324 pKa = 3.52ITGASGTLSAAGQSAVIAGAAVVDD348 pKa = 4.01SDD350 pKa = 5.87FIDD353 pKa = 4.71DD354 pKa = 3.68VLNTVRR360 pKa = 11.84AGATAPPAPTINAVAADD377 pKa = 4.13NIINADD383 pKa = 3.56EE384 pKa = 4.31VEE386 pKa = 4.38SVITGTNEE394 pKa = 3.24TGATVDD400 pKa = 4.51LGIGGNTRR408 pKa = 11.84AATVSGTTWSYY419 pKa = 10.08TLTDD423 pKa = 3.69ADD425 pKa = 4.43LTAMGQGAEE434 pKa = 4.38TLSATQTDD442 pKa = 3.96VLGNTGSAGTRR453 pKa = 11.84AISVDD458 pKa = 3.04TVAPVFSSATVSGRR472 pKa = 11.84TLVLTYY478 pKa = 11.04NEE480 pKa = 4.53ALDD483 pKa = 4.14ATSMPLPSAFRR494 pKa = 11.84VGGIAQGAPVAGVVDD509 pKa = 4.64SAAKK513 pKa = 9.58TLTLTLASPVTYY525 pKa = 9.23GQTGITVSYY534 pKa = 10.26TDD536 pKa = 3.51PTEE539 pKa = 5.28DD540 pKa = 3.55NDD542 pKa = 4.1LAAIQDD548 pKa = 4.04LAGNDD553 pKa = 3.44ASTLTNLAVTNNTPDD568 pKa = 3.2APLYY572 pKa = 10.51QSATVTGSTLVMTYY586 pKa = 10.81AGDD589 pKa = 3.78LDD591 pKa = 4.6EE592 pKa = 6.14NNLPPLDD599 pKa = 3.9AFTVKK604 pKa = 10.4VNNTAQAAPTGVVVDD619 pKa = 4.68ADD621 pKa = 3.82AGTVTLTLAAPVIHH635 pKa = 6.43GQTVTVGYY643 pKa = 8.41TDD645 pKa = 4.36PSAANDD651 pKa = 3.53LNAIQDD657 pKa = 3.62SGLNDD662 pKa = 3.33AVSIGNRR669 pKa = 11.84VVANQTPDD677 pKa = 2.75AAAPVFANAVVTEE690 pKa = 3.91NGKK693 pKa = 9.65YY694 pKa = 10.08LVLNCDD700 pKa = 3.33EE701 pKa = 5.13ALDD704 pKa = 4.29AAHH707 pKa = 6.49VPPAGAFIVKK717 pKa = 10.12VGGVAQATPTGLTVNASAKK736 pKa = 9.72TVMLTLATPITFGQTGITVSYY757 pKa = 9.28TDD759 pKa = 4.08PSAANDD765 pKa = 3.09INAIQDD771 pKa = 3.54AVGNDD776 pKa = 3.44AASVTDD782 pKa = 5.25LPVTNNAADD791 pKa = 3.96TPILEE796 pKa = 4.51SATVHH801 pKa = 6.84GDD803 pKa = 3.68TLVLTYY809 pKa = 11.1NDD811 pKa = 4.46ALDD814 pKa = 4.3AGNVPLPEE822 pKa = 4.26AFAVKK827 pKa = 10.4VGGADD832 pKa = 3.2PVAPTNVAVDD842 pKa = 3.74SAARR846 pKa = 11.84TVTLTLADD854 pKa = 4.03AVTSGQTVKK863 pKa = 10.73LSYY866 pKa = 10.52TDD868 pKa = 3.5PTTDD872 pKa = 3.16NDD874 pKa = 3.24ADD876 pKa = 4.7AIQDD880 pKa = 3.63VAGNDD885 pKa = 3.2AFTVEE890 pKa = 4.36NQSVTNEE897 pKa = 4.06TPPVFSAATVTGSTLVMNYY916 pKa = 10.13SDD918 pKa = 5.06ALDD921 pKa = 4.14AAHH924 pKa = 6.62VPPANAFTIEE934 pKa = 4.39VNGTPQAAPSAVAVDD949 pKa = 3.74AAARR953 pKa = 11.84TVTLTLAAAVTNGQAVTVSYY973 pKa = 11.44ADD975 pKa = 3.45PTAGNDD981 pKa = 3.01AAAIQNAIGSDD992 pKa = 3.47AVSLSNQPVSNHH1004 pKa = 5.48TPPLLSGATVSGDD1017 pKa = 3.65TLVLTYY1023 pKa = 10.97SNEE1026 pKa = 3.82LDD1028 pKa = 3.55EE1029 pKa = 6.02SNIPLVNAFTVKK1041 pKa = 10.55VNGVAQAAPTDD1052 pKa = 4.02VVVDD1056 pKa = 4.13AAARR1060 pKa = 11.84TVTLTLATAVTDD1072 pKa = 4.34GQAVTVSYY1080 pKa = 11.1VDD1082 pKa = 4.05PSTGDD1087 pKa = 3.31DD1088 pKa = 3.47TEE1090 pKa = 5.63AIQDD1094 pKa = 3.91ALGNDD1099 pKa = 4.55AISVANRR1106 pKa = 11.84PVTNTTADD1114 pKa = 3.0VAAPVFSGATVSGDD1128 pKa = 3.6TLVLSYY1134 pKa = 11.24SEE1136 pKa = 5.19ALDD1139 pKa = 3.79AANAPVAGDD1148 pKa = 3.81FIVNVDD1154 pKa = 3.48GTAQADD1160 pKa = 3.68PTAVVVDD1167 pKa = 4.05ATARR1171 pKa = 11.84TVTLTLAAAVAYY1183 pKa = 9.94GEE1185 pKa = 4.55AVTVTYY1191 pKa = 10.24TDD1193 pKa = 3.51PTVGDD1198 pKa = 4.36DD1199 pKa = 3.29ATAIQDD1205 pKa = 3.7TAGNDD1210 pKa = 3.25AVTLTEE1216 pKa = 3.78QSVTNTSVNALAPTLSVVVSAGADD1240 pKa = 3.2VNAIVLTVTTGLNGAASADD1259 pKa = 4.3LIFDD1263 pKa = 4.56GLPGGAVVRR1272 pKa = 11.84NSAAVDD1278 pKa = 3.47VTAGVVNFDD1287 pKa = 3.59GSDD1290 pKa = 3.11TFTVEE1295 pKa = 4.26FPEE1298 pKa = 6.34DD1299 pKa = 3.62SDD1301 pKa = 3.27IHH1303 pKa = 6.72QNLLVTVTGKK1313 pKa = 10.32DD1314 pKa = 3.11AGGDD1318 pKa = 3.58PLGDD1322 pKa = 3.43NVHH1325 pKa = 6.68TIDD1328 pKa = 4.84LVYY1331 pKa = 10.83DD1332 pKa = 3.88VASSTEE1338 pKa = 3.87DD1339 pKa = 3.68LNFASANQNMWGNFNGYY1356 pKa = 9.3IGWHH1360 pKa = 6.07EE1361 pKa = 4.57YY1362 pKa = 10.37IPLLGDD1368 pKa = 4.76APIVWNEE1375 pKa = 3.85ATGEE1379 pKa = 3.92WDD1381 pKa = 3.59DD1382 pKa = 5.31AANPDD1387 pKa = 3.61YY1388 pKa = 10.25WRR1390 pKa = 11.84SGEE1393 pKa = 3.94FSVIDD1398 pKa = 3.5VDD1400 pKa = 4.82LDD1402 pKa = 3.59SSQVIEE1408 pKa = 4.32AAAIAASSVLDD1419 pKa = 3.47TAKK1422 pKa = 10.92AVFDD1426 pKa = 3.94VAAVAVDD1433 pKa = 4.24TVAKK1437 pKa = 10.65GIFDD1441 pKa = 3.54TAKK1444 pKa = 10.77AVFQTAEE1451 pKa = 3.51NLYY1454 pKa = 9.26YY1455 pKa = 10.69FGARR1459 pKa = 11.84TVDD1462 pKa = 3.27AAVQATFQAAKK1473 pKa = 9.76GAYY1476 pKa = 8.98DD1477 pKa = 3.75VAHH1480 pKa = 6.89GLYY1483 pKa = 10.62DD1484 pKa = 3.41SASYY1488 pKa = 10.79LFHH1491 pKa = 6.36TVATGLHH1498 pKa = 5.76DD1499 pKa = 4.56AAWNAYY1505 pKa = 9.52ASYY1508 pKa = 10.81DD1509 pKa = 3.41NWYY1512 pKa = 10.26HH1513 pKa = 7.15SLDD1516 pKa = 3.07GWGQFWNAAGHH1527 pKa = 5.11VANDD1531 pKa = 3.86IAWGIADD1538 pKa = 3.35VAYY1541 pKa = 10.81NVAVPIWDD1549 pKa = 3.45AAKK1552 pKa = 10.58YY1553 pKa = 10.39AFEE1556 pKa = 4.2TTATGIYY1563 pKa = 7.95NTALAVYY1570 pKa = 9.1NAAAGVANSAAQVVFDD1586 pKa = 3.63GAKK1589 pKa = 10.25AVFATAEE1596 pKa = 3.96HH1597 pKa = 6.71VYY1599 pKa = 11.15NEE1601 pKa = 4.33VKK1603 pKa = 10.41QDD1605 pKa = 3.27IYY1607 pKa = 11.73DD1608 pKa = 3.81VAQGVYY1614 pKa = 10.49DD1615 pKa = 4.18LARR1618 pKa = 11.84DD1619 pKa = 3.41GVMAVLATINDD1630 pKa = 3.52KK1631 pKa = 11.51VEE1633 pKa = 3.81FDD1635 pKa = 3.44TALKK1639 pKa = 10.41VDD1641 pKa = 4.17SEE1643 pKa = 4.65VFAQVGLQVDD1653 pKa = 4.94FEE1655 pKa = 5.91LDD1657 pKa = 3.42MGSVDD1662 pKa = 5.51ADD1664 pKa = 2.79IDD1666 pKa = 4.07YY1667 pKa = 11.14QLTSATQYY1675 pKa = 11.32NKK1677 pKa = 10.43AADD1680 pKa = 4.07MLTITPTMTNMTTGDD1695 pKa = 3.47AVAFDD1700 pKa = 4.81TISPNAKK1707 pKa = 9.63FYY1709 pKa = 11.11AALLYY1714 pKa = 10.72DD1715 pKa = 3.75VGADD1719 pKa = 3.19FDD1721 pKa = 4.22VFIDD1725 pKa = 4.02GKK1727 pKa = 10.9LVVDD1731 pKa = 3.59NTTVYY1736 pKa = 10.82DD1737 pKa = 4.62LSPTSDD1743 pKa = 2.99GLAIQFPISTEE1754 pKa = 3.9SFADD1758 pKa = 3.57SLSEE1762 pKa = 3.84MTGGDD1767 pKa = 3.18IEE1769 pKa = 4.36VGKK1772 pKa = 9.5MVLIDD1777 pKa = 4.84LDD1779 pKa = 4.16TTQLEE1784 pKa = 4.39PFEE1787 pKa = 4.59VPFVEE1792 pKa = 6.34ALTQDD1797 pKa = 3.12ILSIEE1802 pKa = 4.06IAIPTLEE1809 pKa = 4.36TEE1811 pKa = 4.88GKK1813 pKa = 9.87AATYY1817 pKa = 10.67VPATADD1823 pKa = 2.51IWTTGTFEE1831 pKa = 4.07EE1832 pKa = 4.82QIGPFPYY1839 pKa = 10.41ASVDD1843 pKa = 3.84FSEE1846 pKa = 4.57ISSAFFNIINAKK1858 pKa = 9.91FDD1860 pKa = 3.85FSEE1863 pKa = 4.47EE1864 pKa = 3.98FMDD1867 pKa = 5.27QYY1869 pKa = 11.94GLDD1872 pKa = 4.0SLGDD1876 pKa = 3.48AATLAEE1882 pKa = 4.49TVQDD1886 pKa = 3.27IATGLMANIWDD1897 pKa = 4.22TLDD1900 pKa = 3.87GEE1902 pKa = 4.67AEE1904 pKa = 4.14GVPIFVLDD1912 pKa = 3.8MTDD1915 pKa = 3.21EE1916 pKa = 4.37TASSLLHH1923 pKa = 6.63LNLFTDD1929 pKa = 4.11DD1930 pKa = 3.69VMGNTTSANTGSLGFYY1946 pKa = 9.86AAYY1949 pKa = 10.25GEE1951 pKa = 4.43SDD1953 pKa = 3.66PVVAVTVDD1961 pKa = 3.7LDD1963 pKa = 3.48AAYY1966 pKa = 10.53VAIVKK1971 pKa = 10.4AIAKK1975 pKa = 9.07AAASAVTAGAATTIHH1990 pKa = 6.38PVIDD1994 pKa = 5.9AIPSPYY2000 pKa = 10.04NLEE2003 pKa = 4.19FGVEE2007 pKa = 4.05QVLEE2011 pKa = 4.19IAAVPQATIDD2021 pKa = 5.64QITQWLNLGFTFEE2034 pKa = 4.71AADD2037 pKa = 4.08LDD2039 pKa = 3.95VTQAVNFSQDD2049 pKa = 3.05FTLSIDD2055 pKa = 3.56DD2056 pKa = 3.57MSYY2059 pKa = 11.29LVTLEE2064 pKa = 5.09DD2065 pKa = 3.7NTTHH2069 pKa = 7.44AFTANGTGALQISNAGSHH2087 pKa = 6.8DD2088 pKa = 4.03ANHH2091 pKa = 7.35DD2092 pKa = 3.58GLITYY2097 pKa = 7.97TLDD2100 pKa = 3.13IVPTAMFSNDD2110 pKa = 3.37TEE2112 pKa = 4.47VGLSLGYY2119 pKa = 10.4QLDD2122 pKa = 3.86FLKK2125 pKa = 11.17GGFSAGAQLPLGEE2138 pKa = 4.92LLGLTDD2144 pKa = 5.76ADD2146 pKa = 3.71WLNIDD2151 pKa = 4.79FSAIDD2156 pKa = 3.41ISMGPLLRR2164 pKa = 11.84VQGEE2168 pKa = 4.79LDD2170 pKa = 3.55TLDD2173 pKa = 3.29VDD2175 pKa = 4.07VFEE2178 pKa = 5.73SRR2180 pKa = 11.84FALDD2184 pKa = 3.31VGSASYY2190 pKa = 10.91AGAIDD2195 pKa = 3.45IGLVNHH2201 pKa = 6.19SVMVV2205 pKa = 3.78
MM1 pKa = 7.7GNFDD5 pKa = 3.99LHH7 pKa = 7.65GLINAIANNHH17 pKa = 5.08GVASPVKK24 pKa = 8.74SAKK27 pKa = 10.04RR28 pKa = 11.84KK29 pKa = 8.43MRR31 pKa = 11.84DD32 pKa = 3.02VSEE35 pKa = 4.82GDD37 pKa = 3.59GQATAQAEE45 pKa = 4.23GDD47 pKa = 3.79ATSATSDD54 pKa = 3.18ASMQLAQADD63 pKa = 3.76TGVATEE69 pKa = 4.24AAAGEE74 pKa = 4.54TTGGASGAPAAGEE87 pKa = 3.84AAAGSGAAAGGLGGLGLGGMALGGAALAGGGGGGGSAVAAAVNNIITGTVVAGPVLAGSGLKK149 pKa = 10.26VEE151 pKa = 5.26IYY153 pKa = 10.3AANGTTKK160 pKa = 10.58LGEE163 pKa = 4.29STLDD167 pKa = 3.2ASGKK171 pKa = 8.51FTVDD175 pKa = 2.53VGAYY179 pKa = 7.71TGIVIARR186 pKa = 11.84LVDD189 pKa = 3.69ANGGDD194 pKa = 4.09DD195 pKa = 4.33YY196 pKa = 11.62LDD198 pKa = 3.91EE199 pKa = 4.51ATGVPKK205 pKa = 10.61DD206 pKa = 3.7LNANLMATSVVTGGTFTLNINPLTTIAAQKK236 pKa = 10.66AGLAADD242 pKa = 4.57GSGGLADD249 pKa = 4.07AAAVANANAAVAAAFGLTDD268 pKa = 3.61LTGTSVVATNGGSYY282 pKa = 10.69DD283 pKa = 3.6AADD286 pKa = 3.51GMSAGEE292 pKa = 4.2KK293 pKa = 10.26YY294 pKa = 10.62GAILAALSGADD305 pKa = 3.73FASSGDD311 pKa = 3.62SQAIIDD317 pKa = 4.25NLVAAIDD324 pKa = 3.52ITGASGTLSAAGQSAVIAGAAVVDD348 pKa = 4.01SDD350 pKa = 5.87FIDD353 pKa = 4.71DD354 pKa = 3.68VLNTVRR360 pKa = 11.84AGATAPPAPTINAVAADD377 pKa = 4.13NIINADD383 pKa = 3.56EE384 pKa = 4.31VEE386 pKa = 4.38SVITGTNEE394 pKa = 3.24TGATVDD400 pKa = 4.51LGIGGNTRR408 pKa = 11.84AATVSGTTWSYY419 pKa = 10.08TLTDD423 pKa = 3.69ADD425 pKa = 4.43LTAMGQGAEE434 pKa = 4.38TLSATQTDD442 pKa = 3.96VLGNTGSAGTRR453 pKa = 11.84AISVDD458 pKa = 3.04TVAPVFSSATVSGRR472 pKa = 11.84TLVLTYY478 pKa = 11.04NEE480 pKa = 4.53ALDD483 pKa = 4.14ATSMPLPSAFRR494 pKa = 11.84VGGIAQGAPVAGVVDD509 pKa = 4.64SAAKK513 pKa = 9.58TLTLTLASPVTYY525 pKa = 9.23GQTGITVSYY534 pKa = 10.26TDD536 pKa = 3.51PTEE539 pKa = 5.28DD540 pKa = 3.55NDD542 pKa = 4.1LAAIQDD548 pKa = 4.04LAGNDD553 pKa = 3.44ASTLTNLAVTNNTPDD568 pKa = 3.2APLYY572 pKa = 10.51QSATVTGSTLVMTYY586 pKa = 10.81AGDD589 pKa = 3.78LDD591 pKa = 4.6EE592 pKa = 6.14NNLPPLDD599 pKa = 3.9AFTVKK604 pKa = 10.4VNNTAQAAPTGVVVDD619 pKa = 4.68ADD621 pKa = 3.82AGTVTLTLAAPVIHH635 pKa = 6.43GQTVTVGYY643 pKa = 8.41TDD645 pKa = 4.36PSAANDD651 pKa = 3.53LNAIQDD657 pKa = 3.62SGLNDD662 pKa = 3.33AVSIGNRR669 pKa = 11.84VVANQTPDD677 pKa = 2.75AAAPVFANAVVTEE690 pKa = 3.91NGKK693 pKa = 9.65YY694 pKa = 10.08LVLNCDD700 pKa = 3.33EE701 pKa = 5.13ALDD704 pKa = 4.29AAHH707 pKa = 6.49VPPAGAFIVKK717 pKa = 10.12VGGVAQATPTGLTVNASAKK736 pKa = 9.72TVMLTLATPITFGQTGITVSYY757 pKa = 9.28TDD759 pKa = 4.08PSAANDD765 pKa = 3.09INAIQDD771 pKa = 3.54AVGNDD776 pKa = 3.44AASVTDD782 pKa = 5.25LPVTNNAADD791 pKa = 3.96TPILEE796 pKa = 4.51SATVHH801 pKa = 6.84GDD803 pKa = 3.68TLVLTYY809 pKa = 11.1NDD811 pKa = 4.46ALDD814 pKa = 4.3AGNVPLPEE822 pKa = 4.26AFAVKK827 pKa = 10.4VGGADD832 pKa = 3.2PVAPTNVAVDD842 pKa = 3.74SAARR846 pKa = 11.84TVTLTLADD854 pKa = 4.03AVTSGQTVKK863 pKa = 10.73LSYY866 pKa = 10.52TDD868 pKa = 3.5PTTDD872 pKa = 3.16NDD874 pKa = 3.24ADD876 pKa = 4.7AIQDD880 pKa = 3.63VAGNDD885 pKa = 3.2AFTVEE890 pKa = 4.36NQSVTNEE897 pKa = 4.06TPPVFSAATVTGSTLVMNYY916 pKa = 10.13SDD918 pKa = 5.06ALDD921 pKa = 4.14AAHH924 pKa = 6.62VPPANAFTIEE934 pKa = 4.39VNGTPQAAPSAVAVDD949 pKa = 3.74AAARR953 pKa = 11.84TVTLTLAAAVTNGQAVTVSYY973 pKa = 11.44ADD975 pKa = 3.45PTAGNDD981 pKa = 3.01AAAIQNAIGSDD992 pKa = 3.47AVSLSNQPVSNHH1004 pKa = 5.48TPPLLSGATVSGDD1017 pKa = 3.65TLVLTYY1023 pKa = 10.97SNEE1026 pKa = 3.82LDD1028 pKa = 3.55EE1029 pKa = 6.02SNIPLVNAFTVKK1041 pKa = 10.55VNGVAQAAPTDD1052 pKa = 4.02VVVDD1056 pKa = 4.13AAARR1060 pKa = 11.84TVTLTLATAVTDD1072 pKa = 4.34GQAVTVSYY1080 pKa = 11.1VDD1082 pKa = 4.05PSTGDD1087 pKa = 3.31DD1088 pKa = 3.47TEE1090 pKa = 5.63AIQDD1094 pKa = 3.91ALGNDD1099 pKa = 4.55AISVANRR1106 pKa = 11.84PVTNTTADD1114 pKa = 3.0VAAPVFSGATVSGDD1128 pKa = 3.6TLVLSYY1134 pKa = 11.24SEE1136 pKa = 5.19ALDD1139 pKa = 3.79AANAPVAGDD1148 pKa = 3.81FIVNVDD1154 pKa = 3.48GTAQADD1160 pKa = 3.68PTAVVVDD1167 pKa = 4.05ATARR1171 pKa = 11.84TVTLTLAAAVAYY1183 pKa = 9.94GEE1185 pKa = 4.55AVTVTYY1191 pKa = 10.24TDD1193 pKa = 3.51PTVGDD1198 pKa = 4.36DD1199 pKa = 3.29ATAIQDD1205 pKa = 3.7TAGNDD1210 pKa = 3.25AVTLTEE1216 pKa = 3.78QSVTNTSVNALAPTLSVVVSAGADD1240 pKa = 3.2VNAIVLTVTTGLNGAASADD1259 pKa = 4.3LIFDD1263 pKa = 4.56GLPGGAVVRR1272 pKa = 11.84NSAAVDD1278 pKa = 3.47VTAGVVNFDD1287 pKa = 3.59GSDD1290 pKa = 3.11TFTVEE1295 pKa = 4.26FPEE1298 pKa = 6.34DD1299 pKa = 3.62SDD1301 pKa = 3.27IHH1303 pKa = 6.72QNLLVTVTGKK1313 pKa = 10.32DD1314 pKa = 3.11AGGDD1318 pKa = 3.58PLGDD1322 pKa = 3.43NVHH1325 pKa = 6.68TIDD1328 pKa = 4.84LVYY1331 pKa = 10.83DD1332 pKa = 3.88VASSTEE1338 pKa = 3.87DD1339 pKa = 3.68LNFASANQNMWGNFNGYY1356 pKa = 9.3IGWHH1360 pKa = 6.07EE1361 pKa = 4.57YY1362 pKa = 10.37IPLLGDD1368 pKa = 4.76APIVWNEE1375 pKa = 3.85ATGEE1379 pKa = 3.92WDD1381 pKa = 3.59DD1382 pKa = 5.31AANPDD1387 pKa = 3.61YY1388 pKa = 10.25WRR1390 pKa = 11.84SGEE1393 pKa = 3.94FSVIDD1398 pKa = 3.5VDD1400 pKa = 4.82LDD1402 pKa = 3.59SSQVIEE1408 pKa = 4.32AAAIAASSVLDD1419 pKa = 3.47TAKK1422 pKa = 10.92AVFDD1426 pKa = 3.94VAAVAVDD1433 pKa = 4.24TVAKK1437 pKa = 10.65GIFDD1441 pKa = 3.54TAKK1444 pKa = 10.77AVFQTAEE1451 pKa = 3.51NLYY1454 pKa = 9.26YY1455 pKa = 10.69FGARR1459 pKa = 11.84TVDD1462 pKa = 3.27AAVQATFQAAKK1473 pKa = 9.76GAYY1476 pKa = 8.98DD1477 pKa = 3.75VAHH1480 pKa = 6.89GLYY1483 pKa = 10.62DD1484 pKa = 3.41SASYY1488 pKa = 10.79LFHH1491 pKa = 6.36TVATGLHH1498 pKa = 5.76DD1499 pKa = 4.56AAWNAYY1505 pKa = 9.52ASYY1508 pKa = 10.81DD1509 pKa = 3.41NWYY1512 pKa = 10.26HH1513 pKa = 7.15SLDD1516 pKa = 3.07GWGQFWNAAGHH1527 pKa = 5.11VANDD1531 pKa = 3.86IAWGIADD1538 pKa = 3.35VAYY1541 pKa = 10.81NVAVPIWDD1549 pKa = 3.45AAKK1552 pKa = 10.58YY1553 pKa = 10.39AFEE1556 pKa = 4.2TTATGIYY1563 pKa = 7.95NTALAVYY1570 pKa = 9.1NAAAGVANSAAQVVFDD1586 pKa = 3.63GAKK1589 pKa = 10.25AVFATAEE1596 pKa = 3.96HH1597 pKa = 6.71VYY1599 pKa = 11.15NEE1601 pKa = 4.33VKK1603 pKa = 10.41QDD1605 pKa = 3.27IYY1607 pKa = 11.73DD1608 pKa = 3.81VAQGVYY1614 pKa = 10.49DD1615 pKa = 4.18LARR1618 pKa = 11.84DD1619 pKa = 3.41GVMAVLATINDD1630 pKa = 3.52KK1631 pKa = 11.51VEE1633 pKa = 3.81FDD1635 pKa = 3.44TALKK1639 pKa = 10.41VDD1641 pKa = 4.17SEE1643 pKa = 4.65VFAQVGLQVDD1653 pKa = 4.94FEE1655 pKa = 5.91LDD1657 pKa = 3.42MGSVDD1662 pKa = 5.51ADD1664 pKa = 2.79IDD1666 pKa = 4.07YY1667 pKa = 11.14QLTSATQYY1675 pKa = 11.32NKK1677 pKa = 10.43AADD1680 pKa = 4.07MLTITPTMTNMTTGDD1695 pKa = 3.47AVAFDD1700 pKa = 4.81TISPNAKK1707 pKa = 9.63FYY1709 pKa = 11.11AALLYY1714 pKa = 10.72DD1715 pKa = 3.75VGADD1719 pKa = 3.19FDD1721 pKa = 4.22VFIDD1725 pKa = 4.02GKK1727 pKa = 10.9LVVDD1731 pKa = 3.59NTTVYY1736 pKa = 10.82DD1737 pKa = 4.62LSPTSDD1743 pKa = 2.99GLAIQFPISTEE1754 pKa = 3.9SFADD1758 pKa = 3.57SLSEE1762 pKa = 3.84MTGGDD1767 pKa = 3.18IEE1769 pKa = 4.36VGKK1772 pKa = 9.5MVLIDD1777 pKa = 4.84LDD1779 pKa = 4.16TTQLEE1784 pKa = 4.39PFEE1787 pKa = 4.59VPFVEE1792 pKa = 6.34ALTQDD1797 pKa = 3.12ILSIEE1802 pKa = 4.06IAIPTLEE1809 pKa = 4.36TEE1811 pKa = 4.88GKK1813 pKa = 9.87AATYY1817 pKa = 10.67VPATADD1823 pKa = 2.51IWTTGTFEE1831 pKa = 4.07EE1832 pKa = 4.82QIGPFPYY1839 pKa = 10.41ASVDD1843 pKa = 3.84FSEE1846 pKa = 4.57ISSAFFNIINAKK1858 pKa = 9.91FDD1860 pKa = 3.85FSEE1863 pKa = 4.47EE1864 pKa = 3.98FMDD1867 pKa = 5.27QYY1869 pKa = 11.94GLDD1872 pKa = 4.0SLGDD1876 pKa = 3.48AATLAEE1882 pKa = 4.49TVQDD1886 pKa = 3.27IATGLMANIWDD1897 pKa = 4.22TLDD1900 pKa = 3.87GEE1902 pKa = 4.67AEE1904 pKa = 4.14GVPIFVLDD1912 pKa = 3.8MTDD1915 pKa = 3.21EE1916 pKa = 4.37TASSLLHH1923 pKa = 6.63LNLFTDD1929 pKa = 4.11DD1930 pKa = 3.69VMGNTTSANTGSLGFYY1946 pKa = 9.86AAYY1949 pKa = 10.25GEE1951 pKa = 4.43SDD1953 pKa = 3.66PVVAVTVDD1961 pKa = 3.7LDD1963 pKa = 3.48AAYY1966 pKa = 10.53VAIVKK1971 pKa = 10.4AIAKK1975 pKa = 9.07AAASAVTAGAATTIHH1990 pKa = 6.38PVIDD1994 pKa = 5.9AIPSPYY2000 pKa = 10.04NLEE2003 pKa = 4.19FGVEE2007 pKa = 4.05QVLEE2011 pKa = 4.19IAAVPQATIDD2021 pKa = 5.64QITQWLNLGFTFEE2034 pKa = 4.71AADD2037 pKa = 4.08LDD2039 pKa = 3.95VTQAVNFSQDD2049 pKa = 3.05FTLSIDD2055 pKa = 3.56DD2056 pKa = 3.57MSYY2059 pKa = 11.29LVTLEE2064 pKa = 5.09DD2065 pKa = 3.7NTTHH2069 pKa = 7.44AFTANGTGALQISNAGSHH2087 pKa = 6.8DD2088 pKa = 4.03ANHH2091 pKa = 7.35DD2092 pKa = 3.58GLITYY2097 pKa = 7.97TLDD2100 pKa = 3.13IVPTAMFSNDD2110 pKa = 3.37TEE2112 pKa = 4.47VGLSLGYY2119 pKa = 10.4QLDD2122 pKa = 3.86FLKK2125 pKa = 11.17GGFSAGAQLPLGEE2138 pKa = 4.92LLGLTDD2144 pKa = 5.76ADD2146 pKa = 3.71WLNIDD2151 pKa = 4.79FSAIDD2156 pKa = 3.41ISMGPLLRR2164 pKa = 11.84VQGEE2168 pKa = 4.79LDD2170 pKa = 3.55TLDD2173 pKa = 3.29VDD2175 pKa = 4.07VFEE2178 pKa = 5.73SRR2180 pKa = 11.84FALDD2184 pKa = 3.31VGSASYY2190 pKa = 10.91AGAIDD2195 pKa = 3.45IGLVNHH2201 pKa = 6.19SVMVV2205 pKa = 3.78
Molecular weight: 223.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0SL05|W0SL05_9PROT PAS/PAC sensor-containing diguanylate cyclase/phosphodiesterase OS=Sulfuritalea hydrogenivorans sk43H OX=1223802 GN=SUTH_02684 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.72GFLVRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.6RR14 pKa = 11.84THH16 pKa = 5.72GFLVRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84GGRR28 pKa = 11.84AVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.91GRR39 pKa = 11.84HH40 pKa = 4.92RR41 pKa = 11.84LAVV44 pKa = 3.37
Molecular weight: 5.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1166199 |
39 |
4918 |
325.9 |
35.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.399 ± 0.067 | 1.01 ± 0.017 |
5.375 ± 0.034 | 5.82 ± 0.041 |
3.668 ± 0.031 | 8.262 ± 0.054 |
2.227 ± 0.021 | 5.03 ± 0.031 |
3.917 ± 0.042 | 10.654 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.544 ± 0.02 | 2.785 ± 0.03 |
4.888 ± 0.033 | 3.469 ± 0.04 |
6.866 ± 0.055 | 5.192 ± 0.037 |
5.029 ± 0.072 | 7.242 ± 0.036 |
1.36 ± 0.018 | 2.262 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
