
Methanobacterium virus PhiF1
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
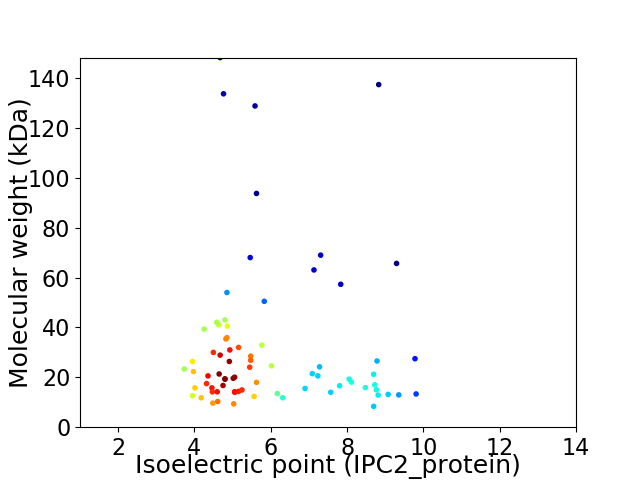
Virtual 2D-PAGE plot for 76 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W642|A0A5Q2W642_9VIRU Putative glycosyltransferase OS=Methanobacterium virus PhiF1 OX=2609788 PE=4 SV=1
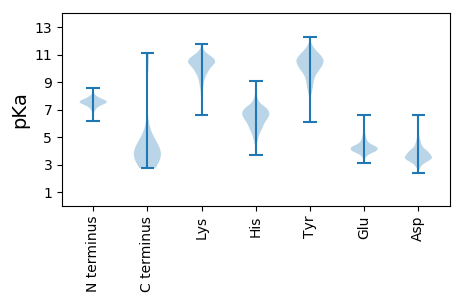
MM1 pKa = 7.43VYY3 pKa = 10.92VMWLDD8 pKa = 3.27GRR10 pKa = 11.84PIGIRR15 pKa = 11.84DD16 pKa = 3.52DD17 pKa = 3.97EE18 pKa = 4.58FPGFYY23 pKa = 10.57EE24 pKa = 4.1EE25 pKa = 4.77TVDD28 pKa = 3.65EE29 pKa = 5.22ALIEE33 pKa = 4.37DD34 pKa = 4.29LMGVNPDD41 pKa = 2.93NMRR44 pKa = 11.84LVGYY48 pKa = 10.5FEE50 pKa = 5.42DD51 pKa = 4.96RR52 pKa = 11.84EE53 pKa = 4.23ALEE56 pKa = 4.16EE57 pKa = 4.07AVAPEE62 pKa = 4.72DD63 pKa = 3.68PDD65 pKa = 3.85MIIWDD70 pKa = 4.03GPDD73 pKa = 3.35FYY75 pKa = 11.02LYY77 pKa = 10.0WGVPDD82 pKa = 5.64PIVATWRR89 pKa = 11.84YY90 pKa = 9.4VLDD93 pKa = 4.26FEE95 pKa = 6.03VDD97 pKa = 3.63YY98 pKa = 11.37QDD100 pKa = 4.81YY101 pKa = 10.53EE102 pKa = 4.41VAQALSDD109 pKa = 3.48FRR111 pKa = 11.84DD112 pKa = 3.55YY113 pKa = 11.68VDD115 pKa = 4.23YY116 pKa = 11.36LEE118 pKa = 4.31QPGVISFEE126 pKa = 4.22EE127 pKa = 4.29EE128 pKa = 3.83TVLRR132 pKa = 11.84NLRR135 pKa = 11.84DD136 pKa = 3.56QLIDD140 pKa = 3.23SHH142 pKa = 5.52THH144 pKa = 4.66MEE146 pKa = 3.89VDD148 pKa = 3.71EE149 pKa = 4.2VLEE152 pKa = 3.91ALIDD156 pKa = 3.83RR157 pKa = 11.84LEE159 pKa = 5.02DD160 pKa = 3.13ICGDD164 pKa = 3.33RR165 pKa = 11.84LYY167 pKa = 10.61WVIDD171 pKa = 3.65PLCNGEE177 pKa = 6.26LKK179 pKa = 8.85TQWHH183 pKa = 6.2VDD185 pKa = 3.56LEE187 pKa = 4.36QEE189 pKa = 4.16LLYY192 pKa = 10.78KK193 pKa = 10.58VEE195 pKa = 4.26GLNN198 pKa = 4.06
MM1 pKa = 7.43VYY3 pKa = 10.92VMWLDD8 pKa = 3.27GRR10 pKa = 11.84PIGIRR15 pKa = 11.84DD16 pKa = 3.52DD17 pKa = 3.97EE18 pKa = 4.58FPGFYY23 pKa = 10.57EE24 pKa = 4.1EE25 pKa = 4.77TVDD28 pKa = 3.65EE29 pKa = 5.22ALIEE33 pKa = 4.37DD34 pKa = 4.29LMGVNPDD41 pKa = 2.93NMRR44 pKa = 11.84LVGYY48 pKa = 10.5FEE50 pKa = 5.42DD51 pKa = 4.96RR52 pKa = 11.84EE53 pKa = 4.23ALEE56 pKa = 4.16EE57 pKa = 4.07AVAPEE62 pKa = 4.72DD63 pKa = 3.68PDD65 pKa = 3.85MIIWDD70 pKa = 4.03GPDD73 pKa = 3.35FYY75 pKa = 11.02LYY77 pKa = 10.0WGVPDD82 pKa = 5.64PIVATWRR89 pKa = 11.84YY90 pKa = 9.4VLDD93 pKa = 4.26FEE95 pKa = 6.03VDD97 pKa = 3.63YY98 pKa = 11.37QDD100 pKa = 4.81YY101 pKa = 10.53EE102 pKa = 4.41VAQALSDD109 pKa = 3.48FRR111 pKa = 11.84DD112 pKa = 3.55YY113 pKa = 11.68VDD115 pKa = 4.23YY116 pKa = 11.36LEE118 pKa = 4.31QPGVISFEE126 pKa = 4.22EE127 pKa = 4.29EE128 pKa = 3.83TVLRR132 pKa = 11.84NLRR135 pKa = 11.84DD136 pKa = 3.56QLIDD140 pKa = 3.23SHH142 pKa = 5.52THH144 pKa = 4.66MEE146 pKa = 3.89VDD148 pKa = 3.71EE149 pKa = 4.2VLEE152 pKa = 3.91ALIDD156 pKa = 3.83RR157 pKa = 11.84LEE159 pKa = 5.02DD160 pKa = 3.13ICGDD164 pKa = 3.33RR165 pKa = 11.84LYY167 pKa = 10.61WVIDD171 pKa = 3.65PLCNGEE177 pKa = 6.26LKK179 pKa = 8.85TQWHH183 pKa = 6.2VDD185 pKa = 3.56LEE187 pKa = 4.36QEE189 pKa = 4.16LLYY192 pKa = 10.78KK193 pKa = 10.58VEE195 pKa = 4.26GLNN198 pKa = 4.06
Molecular weight: 23.33 kDa
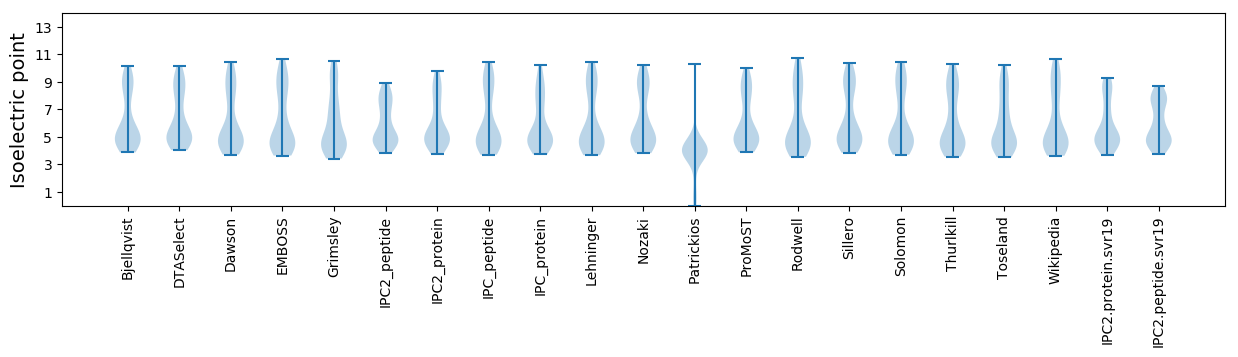
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W4S8|A0A5Q2W4S8_9VIRU Uncharacterized protein OS=Methanobacterium virus PhiF1 OX=2609788 PE=4 SV=1
MM1 pKa = 7.69LEE3 pKa = 3.97LAVNFAQAEE12 pKa = 4.25KK13 pKa = 10.34QGRR16 pKa = 11.84IVLANSYY23 pKa = 11.18AEE25 pKa = 4.38AEE27 pKa = 4.21FLEE30 pKa = 4.44RR31 pKa = 11.84TLPEE35 pKa = 3.93KK36 pKa = 10.3AAEE39 pKa = 3.95MLAAYY44 pKa = 8.78IQYY47 pKa = 10.3HH48 pKa = 5.99ILHH51 pKa = 6.38NQTKK55 pKa = 9.63GAPLYY60 pKa = 10.67EE61 pKa = 4.53SGNLYY66 pKa = 10.51NSVYY70 pKa = 9.99IDD72 pKa = 3.27QVGANDD78 pKa = 3.74YY79 pKa = 9.93MVGVYY84 pKa = 10.6ALDD87 pKa = 3.59EE88 pKa = 4.49KK89 pKa = 11.16GRR91 pKa = 11.84DD92 pKa = 3.44YY93 pKa = 11.41AIVLEE98 pKa = 4.23YY99 pKa = 11.2GLGNLKK105 pKa = 10.43GSYY108 pKa = 10.11PFFRR112 pKa = 11.84PALQQFFFDD121 pKa = 3.78VDD123 pKa = 4.13NITSLLTNSRR133 pKa = 11.84SPALPPLKK141 pKa = 10.56DD142 pKa = 2.99RR143 pKa = 11.84ASPIRR148 pKa = 11.84RR149 pKa = 11.84AAAYY153 pKa = 9.38KK154 pKa = 9.98GANKK158 pKa = 9.93RR159 pKa = 11.84RR160 pKa = 11.84MKK162 pKa = 10.28KK163 pKa = 9.38ISRR166 pKa = 11.84IKK168 pKa = 8.91GGYY171 pKa = 7.66YY172 pKa = 8.52RR173 pKa = 11.84WHH175 pKa = 6.23GRR177 pKa = 11.84RR178 pKa = 11.84EE179 pKa = 3.94EE180 pKa = 3.84FRR182 pKa = 11.84LYY184 pKa = 9.66LTSKK188 pKa = 9.94FGGRR192 pKa = 11.84FGTRR196 pKa = 11.84RR197 pKa = 11.84PEE199 pKa = 3.95AMPMGTKK206 pKa = 9.88RR207 pKa = 11.84QAYY210 pKa = 9.13RR211 pKa = 11.84LAKK214 pKa = 9.93GRR216 pKa = 11.84AASGRR221 pKa = 11.84PKK223 pKa = 10.03TISKK227 pKa = 9.82KK228 pKa = 10.17RR229 pKa = 11.84PEE231 pKa = 4.34GYY233 pKa = 9.79LRR235 pKa = 11.84HH236 pKa = 6.25KK237 pKa = 10.75RR238 pKa = 11.84RR239 pKa = 11.84DD240 pKa = 3.23
MM1 pKa = 7.69LEE3 pKa = 3.97LAVNFAQAEE12 pKa = 4.25KK13 pKa = 10.34QGRR16 pKa = 11.84IVLANSYY23 pKa = 11.18AEE25 pKa = 4.38AEE27 pKa = 4.21FLEE30 pKa = 4.44RR31 pKa = 11.84TLPEE35 pKa = 3.93KK36 pKa = 10.3AAEE39 pKa = 3.95MLAAYY44 pKa = 8.78IQYY47 pKa = 10.3HH48 pKa = 5.99ILHH51 pKa = 6.38NQTKK55 pKa = 9.63GAPLYY60 pKa = 10.67EE61 pKa = 4.53SGNLYY66 pKa = 10.51NSVYY70 pKa = 9.99IDD72 pKa = 3.27QVGANDD78 pKa = 3.74YY79 pKa = 9.93MVGVYY84 pKa = 10.6ALDD87 pKa = 3.59EE88 pKa = 4.49KK89 pKa = 11.16GRR91 pKa = 11.84DD92 pKa = 3.44YY93 pKa = 11.41AIVLEE98 pKa = 4.23YY99 pKa = 11.2GLGNLKK105 pKa = 10.43GSYY108 pKa = 10.11PFFRR112 pKa = 11.84PALQQFFFDD121 pKa = 3.78VDD123 pKa = 4.13NITSLLTNSRR133 pKa = 11.84SPALPPLKK141 pKa = 10.56DD142 pKa = 2.99RR143 pKa = 11.84ASPIRR148 pKa = 11.84RR149 pKa = 11.84AAAYY153 pKa = 9.38KK154 pKa = 9.98GANKK158 pKa = 9.93RR159 pKa = 11.84RR160 pKa = 11.84MKK162 pKa = 10.28KK163 pKa = 9.38ISRR166 pKa = 11.84IKK168 pKa = 8.91GGYY171 pKa = 7.66YY172 pKa = 8.52RR173 pKa = 11.84WHH175 pKa = 6.23GRR177 pKa = 11.84RR178 pKa = 11.84EE179 pKa = 3.94EE180 pKa = 3.84FRR182 pKa = 11.84LYY184 pKa = 9.66LTSKK188 pKa = 9.94FGGRR192 pKa = 11.84FGTRR196 pKa = 11.84RR197 pKa = 11.84PEE199 pKa = 3.95AMPMGTKK206 pKa = 9.88RR207 pKa = 11.84QAYY210 pKa = 9.13RR211 pKa = 11.84LAKK214 pKa = 9.93GRR216 pKa = 11.84AASGRR221 pKa = 11.84PKK223 pKa = 10.03TISKK227 pKa = 9.82KK228 pKa = 10.17RR229 pKa = 11.84PEE231 pKa = 4.34GYY233 pKa = 9.79LRR235 pKa = 11.84HH236 pKa = 6.25KK237 pKa = 10.75RR238 pKa = 11.84RR239 pKa = 11.84DD240 pKa = 3.23
Molecular weight: 27.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
21428 |
76 |
1310 |
281.9 |
31.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.805 ± 0.383 | 1.265 ± 0.159 |
6.613 ± 0.278 | 7.07 ± 0.255 |
3.892 ± 0.212 | 7.705 ± 0.328 |
1.652 ± 0.137 | 7.187 ± 0.275 |
5.432 ± 0.305 | 7.938 ± 0.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.627 ± 0.169 | 3.985 ± 0.224 |
4.732 ± 0.204 | 2.632 ± 0.217 |
5.843 ± 0.277 | 6.258 ± 0.263 |
6.249 ± 0.573 | 6.818 ± 0.397 |
1.213 ± 0.107 | 5.082 ± 0.282 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
