
Sulfurospirillum cavolei
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Campylobacteraceae; Sulfurospirillum
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
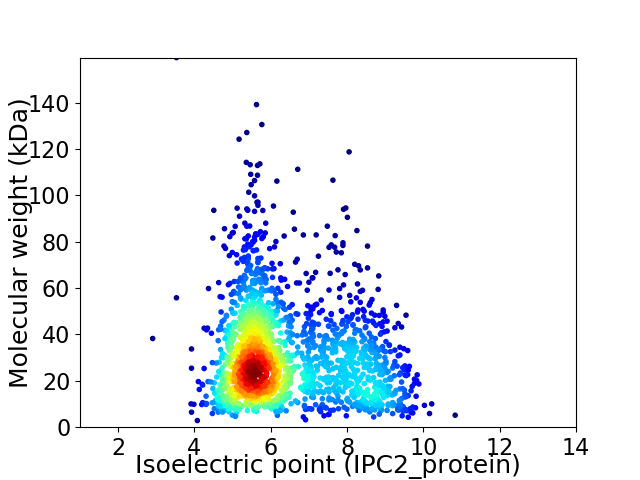
Virtual 2D-PAGE plot for 1874 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D3WDG9|A0A2D3WDG9_9PROT Nitrogenase molybdenum-cofactor biosynthesis protein NifE OS=Sulfurospirillum cavolei OX=366522 GN=CFH80_01145 PE=3 SV=1
MM1 pKa = 7.46SNAPEE6 pKa = 4.15VIYY9 pKa = 10.9SKK11 pKa = 10.02STISLKK17 pKa = 10.65SLPAKK22 pKa = 10.51GEE24 pKa = 3.89NLNLFLRR31 pKa = 11.84PGDD34 pKa = 4.08EE35 pKa = 4.68LSLALDD41 pKa = 3.82LSKK44 pKa = 11.41AKK46 pKa = 10.29FQVVGGDD53 pKa = 4.01IIATLPSGGQLTFVSLGMMAFEE75 pKa = 4.83NNAPVIKK82 pKa = 10.38LPNGTFLALEE92 pKa = 4.36QILDD96 pKa = 4.06TITDD100 pKa = 3.57VGQATKK106 pKa = 10.82DD107 pKa = 3.37AVLVSGNVSLEE118 pKa = 4.13TQEE121 pKa = 4.32QPIEE125 pKa = 4.15HH126 pKa = 5.86EE127 pKa = 4.51QKK129 pKa = 11.08AKK131 pKa = 9.36TQDD134 pKa = 3.39APVNDD139 pKa = 3.62YY140 pKa = 10.52NAYY143 pKa = 10.14YY144 pKa = 10.55VDD146 pKa = 3.91PEE148 pKa = 4.29LPKK151 pKa = 10.8RR152 pKa = 11.84LDD154 pKa = 3.61DD155 pKa = 3.79VASKK159 pKa = 11.11DD160 pKa = 3.59EE161 pKa = 4.27SGKK164 pKa = 10.22YY165 pKa = 9.25LQEE168 pKa = 4.18SLPEE172 pKa = 3.99FTSNEE177 pKa = 4.05TAKK180 pKa = 10.85EE181 pKa = 3.89DD182 pKa = 3.62LTNKK186 pKa = 9.89YY187 pKa = 10.31SSSEE191 pKa = 4.08EE192 pKa = 3.84KK193 pKa = 10.37TKK195 pKa = 11.48DD196 pKa = 3.21NVADD200 pKa = 3.69VSAALSFDD208 pKa = 3.28VGFYY212 pKa = 9.93QIKK215 pKa = 10.22GSEE218 pKa = 4.14STADD222 pKa = 3.14SVTSVLGGTGSALGNVSKK240 pKa = 10.81LAAAQFEE247 pKa = 4.91PEE249 pKa = 3.97TLDD252 pKa = 3.54YY253 pKa = 11.18RR254 pKa = 11.84NSAYY258 pKa = 9.75STVITADD265 pKa = 2.89NPAFVNATYY274 pKa = 8.72LTKK277 pKa = 10.53LVRR280 pKa = 11.84LNVSQPIGFTITSIAINGLTSGGFEE305 pKa = 4.13ILNSDD310 pKa = 4.59FSSAGNASGGWTLSAGTGFSSTLSDD335 pKa = 3.13GGEE338 pKa = 4.0TIEE341 pKa = 5.3FYY343 pKa = 10.54IRR345 pKa = 11.84YY346 pKa = 9.32YY347 pKa = 9.81PXNFDD352 pKa = 3.96NLMQVALTSEE362 pKa = 4.67FNMSNVPEE370 pKa = 4.26EE371 pKa = 4.13KK372 pKa = 10.18QGDD375 pKa = 4.34VIVPDD380 pKa = 3.79LTKK383 pKa = 10.06LTAYY387 pKa = 9.84KK388 pKa = 10.49DD389 pKa = 2.8IGVIVKK395 pKa = 9.98EE396 pKa = 3.96VDD398 pKa = 3.41SVSDD402 pKa = 3.33YY403 pKa = 11.0TYY405 pKa = 10.4TGKK408 pKa = 10.15HH409 pKa = 3.99STGFVLDD416 pKa = 3.61TTPNEE421 pKa = 4.27NIVYY425 pKa = 9.12TSKK428 pKa = 11.08LDD430 pKa = 3.56STVTGGLSNDD440 pKa = 3.18TMYY443 pKa = 11.48GSIGNDD449 pKa = 3.0TLEE452 pKa = 4.48GSNGNDD458 pKa = 3.39TLSGGAGVNTLIGGSGIDD476 pKa = 3.2TADD479 pKa = 3.18YY480 pKa = 11.46SFVTKK485 pKa = 10.88YY486 pKa = 11.1SDD488 pKa = 4.42DD489 pKa = 3.72FLFANDD495 pKa = 3.52GHH497 pKa = 6.94IDD499 pKa = 3.68PVTGVAYY506 pKa = 10.35ANDD509 pKa = 3.91SKK511 pKa = 11.64GVIVDD516 pKa = 4.25LQAGAASGKK525 pKa = 7.32TVYY528 pKa = 10.56DD529 pKa = 3.39ATSDD533 pKa = 3.66VLTGEE538 pKa = 4.3EE539 pKa = 4.11SRR541 pKa = 11.84TISDD545 pKa = 3.52HH546 pKa = 6.69LSGIEE551 pKa = 3.75YY552 pKa = 10.66VVGSKK557 pKa = 10.84YY558 pKa = 10.93DD559 pKa = 3.34DD560 pKa = 4.28TIRR563 pKa = 11.84GDD565 pKa = 3.46ANANKK570 pKa = 10.35LSGGEE575 pKa = 4.2GNDD578 pKa = 3.53TLEE581 pKa = 4.68GRR583 pKa = 11.84DD584 pKa = 3.54GKK586 pKa = 10.71DD587 pKa = 2.77WLEE590 pKa = 4.27GGAGNDD596 pKa = 3.41WLIGSTDD603 pKa = 3.78DD604 pKa = 4.19YY605 pKa = 11.13MIDD608 pKa = 4.1GGDD611 pKa = 3.63NTDD614 pKa = 3.58TADD617 pKa = 4.3FSNNSNGLIITLNDD631 pKa = 3.45SANGSLGNIGGTSATTIIKK650 pKa = 10.21NVEE653 pKa = 4.06NVAGTQYY660 pKa = 11.09KK661 pKa = 10.69DD662 pKa = 3.69SISGDD667 pKa = 3.45TADD670 pKa = 3.99NLLIGGYY677 pKa = 8.44DD678 pKa = 3.41HH679 pKa = 7.47SATAVTANDD688 pKa = 3.51DD689 pKa = 4.28TITGGSGADD698 pKa = 3.56TIVGDD703 pKa = 3.79MMIDD707 pKa = 3.54TVIATPLYY715 pKa = 10.59AGNDD719 pKa = 3.67VLGGGSDD726 pKa = 3.51NDD728 pKa = 4.3TIYY731 pKa = 11.34GDD733 pKa = 3.78SAPILSNAEE742 pKa = 3.82VEE744 pKa = 4.64VAADD748 pKa = 3.34QSVEE752 pKa = 4.22YY753 pKa = 10.36IRR755 pKa = 11.84NTDD758 pKa = 3.85DD759 pKa = 5.19DD760 pKa = 4.25STLATIYY767 pKa = 10.76GGNDD771 pKa = 3.16TLKK774 pKa = 11.09GGGGDD779 pKa = 3.93DD780 pKa = 3.89YY781 pKa = 11.87LNGGSGWDD789 pKa = 3.5TVDD792 pKa = 3.7YY793 pKa = 10.16SASAAAVIVNLSSNYY808 pKa = 10.77AMGEE812 pKa = 4.29GYY814 pKa = 10.29DD815 pKa = 3.27QLFNIEE821 pKa = 4.82NIVGSNSALGDD832 pKa = 3.76TLIGNALANTIVGSTSGADD851 pKa = 3.46TLSGMGGLDD860 pKa = 3.38TLNGLGGKK868 pKa = 9.16DD869 pKa = 3.01WADD872 pKa = 3.31YY873 pKa = 10.79SYY875 pKa = 11.96VSGTTGVNVDD885 pKa = 3.95LTAGTAYY892 pKa = 10.62VSATDD897 pKa = 3.85GDD899 pKa = 4.01TLIAIEE905 pKa = 4.11NVIGSSKK912 pKa = 11.18ADD914 pKa = 3.66TLSGGSGNSVNTLMGGLGNDD934 pKa = 2.7IFYY937 pKa = 10.54GYY939 pKa = 11.14GDD941 pKa = 4.31GDD943 pKa = 4.29LLDD946 pKa = 4.96GGSGSDD952 pKa = 3.19TADD955 pKa = 3.03YY956 pKa = 9.17MYY958 pKa = 10.8AQGRR962 pKa = 11.84IVVTLDD968 pKa = 3.22GSNSDD973 pKa = 3.01TLMNIEE979 pKa = 4.19NVYY982 pKa = 10.26GASDD986 pKa = 3.95KK987 pKa = 10.63DD988 pKa = 3.81TLIGNSDD995 pKa = 3.54ANILDD1000 pKa = 3.56GRR1002 pKa = 11.84ADD1004 pKa = 3.79TDD1006 pKa = 3.78TLSGMGGADD1015 pKa = 3.53TLIGGAGDD1023 pKa = 3.53DD1024 pKa = 4.13TLIGGAGNDD1033 pKa = 4.2TLWGGTDD1040 pKa = 3.24ATHH1043 pKa = 8.06DD1044 pKa = 3.98SGIDD1048 pKa = 3.18TADD1051 pKa = 3.4YY1052 pKa = 10.62SGSSSIYY1059 pKa = 10.32ADD1061 pKa = 3.66LANGTVNDD1069 pKa = 4.43GLGGIDD1075 pKa = 3.34TLAGIEE1081 pKa = 4.34VILGSGSNDD1090 pKa = 3.15VMIGSGNADD1099 pKa = 3.46TLIGNGGNDD1108 pKa = 3.16TFFASGGGDD1117 pKa = 3.04SYY1119 pKa = 11.78YY1120 pKa = 11.16GGVFGSSSDD1129 pKa = 3.65GNVKK1133 pKa = 10.46DD1134 pKa = 3.74RR1135 pKa = 11.84LSYY1138 pKa = 11.09ADD1140 pKa = 4.04VYY1142 pKa = 11.34HH1143 pKa = 7.34DD1144 pKa = 4.61VIDD1147 pKa = 4.27SKK1149 pKa = 10.07TVDD1152 pKa = 4.19HH1153 pKa = 6.67IVVDD1157 pKa = 5.01LSHH1160 pKa = 6.92VGTNATILDD1169 pKa = 3.76ASNAVISTDD1178 pKa = 2.83SLYY1181 pKa = 10.84AIEE1184 pKa = 5.43EE1185 pKa = 4.16IVATDD1190 pKa = 3.64GNDD1193 pKa = 3.52TLRR1196 pKa = 11.84GGAGSQQTFYY1206 pKa = 11.34GGAGDD1211 pKa = 4.54DD1212 pKa = 3.87TLSGNTDD1219 pKa = 2.74GDD1221 pKa = 4.11YY1222 pKa = 11.5LXTNLADD1229 pKa = 3.58YY1230 pKa = 7.63TAQSSNLVVNLASTSNNVYY1249 pKa = 10.65LSGSSPTSSNSDD1261 pKa = 2.93TLANINNIATGSGADD1276 pKa = 3.63TIIGNGNDD1284 pKa = 3.53NIVRR1288 pKa = 11.84AGVGADD1294 pKa = 3.62TLSGGAGDD1302 pKa = 3.63DD1303 pKa = 3.69TLYY1306 pKa = 11.52GEE1308 pKa = 5.21AGNDD1312 pKa = 3.54TLSGGEE1318 pKa = 4.39GNDD1321 pKa = 3.55TLIGGMSTSVDD1332 pKa = 3.57SGIDD1336 pKa = 3.28TADD1339 pKa = 3.47YY1340 pKa = 11.04SATSYY1345 pKa = 11.43YY1346 pKa = 10.69GVTVDD1351 pKa = 4.72LQNGNATGATIGTDD1365 pKa = 2.84KK1366 pKa = 11.15LYY1368 pKa = 11.02GIEE1371 pKa = 4.11NVLGSAQSDD1380 pKa = 3.92TLIGASGTLNTLSGLGGNDD1399 pKa = 2.99TLYY1402 pKa = 11.53GNLEE1406 pKa = 3.76GDD1408 pKa = 4.04YY1409 pKa = 10.95LDD1411 pKa = 5.23GGSGTNTADD1420 pKa = 3.6YY1421 pKa = 10.85SSEE1424 pKa = 4.02TASIVANLTTGRR1436 pKa = 11.84AYY1438 pKa = 8.58FTSSSSYY1445 pKa = 10.66DD1446 pKa = 3.37ALVSIQNVLSGSGNDD1461 pKa = 3.65TLIGLSGTLNTLNGNDD1477 pKa = 4.39GDD1479 pKa = 4.36DD1480 pKa = 3.84TLSGNLDD1487 pKa = 3.58GDD1489 pKa = 4.21SLIGGDD1495 pKa = 4.72GSDD1498 pKa = 3.38TLDD1501 pKa = 3.35YY1502 pKa = 7.98TTANAALTVDD1512 pKa = 5.08LGTQTLFKK1520 pKa = 10.38SASSSAKK1527 pKa = 10.04DD1528 pKa = 3.71YY1529 pKa = 10.89YY1530 pKa = 11.29SEE1532 pKa = 5.19AEE1534 pKa = 4.3VIVTGTYY1541 pKa = 10.33NDD1543 pKa = 3.82SFTLSTSTDD1552 pKa = 3.37TAAMTLDD1559 pKa = 3.83GGAGG1563 pKa = 3.22
MM1 pKa = 7.46SNAPEE6 pKa = 4.15VIYY9 pKa = 10.9SKK11 pKa = 10.02STISLKK17 pKa = 10.65SLPAKK22 pKa = 10.51GEE24 pKa = 3.89NLNLFLRR31 pKa = 11.84PGDD34 pKa = 4.08EE35 pKa = 4.68LSLALDD41 pKa = 3.82LSKK44 pKa = 11.41AKK46 pKa = 10.29FQVVGGDD53 pKa = 4.01IIATLPSGGQLTFVSLGMMAFEE75 pKa = 4.83NNAPVIKK82 pKa = 10.38LPNGTFLALEE92 pKa = 4.36QILDD96 pKa = 4.06TITDD100 pKa = 3.57VGQATKK106 pKa = 10.82DD107 pKa = 3.37AVLVSGNVSLEE118 pKa = 4.13TQEE121 pKa = 4.32QPIEE125 pKa = 4.15HH126 pKa = 5.86EE127 pKa = 4.51QKK129 pKa = 11.08AKK131 pKa = 9.36TQDD134 pKa = 3.39APVNDD139 pKa = 3.62YY140 pKa = 10.52NAYY143 pKa = 10.14YY144 pKa = 10.55VDD146 pKa = 3.91PEE148 pKa = 4.29LPKK151 pKa = 10.8RR152 pKa = 11.84LDD154 pKa = 3.61DD155 pKa = 3.79VASKK159 pKa = 11.11DD160 pKa = 3.59EE161 pKa = 4.27SGKK164 pKa = 10.22YY165 pKa = 9.25LQEE168 pKa = 4.18SLPEE172 pKa = 3.99FTSNEE177 pKa = 4.05TAKK180 pKa = 10.85EE181 pKa = 3.89DD182 pKa = 3.62LTNKK186 pKa = 9.89YY187 pKa = 10.31SSSEE191 pKa = 4.08EE192 pKa = 3.84KK193 pKa = 10.37TKK195 pKa = 11.48DD196 pKa = 3.21NVADD200 pKa = 3.69VSAALSFDD208 pKa = 3.28VGFYY212 pKa = 9.93QIKK215 pKa = 10.22GSEE218 pKa = 4.14STADD222 pKa = 3.14SVTSVLGGTGSALGNVSKK240 pKa = 10.81LAAAQFEE247 pKa = 4.91PEE249 pKa = 3.97TLDD252 pKa = 3.54YY253 pKa = 11.18RR254 pKa = 11.84NSAYY258 pKa = 9.75STVITADD265 pKa = 2.89NPAFVNATYY274 pKa = 8.72LTKK277 pKa = 10.53LVRR280 pKa = 11.84LNVSQPIGFTITSIAINGLTSGGFEE305 pKa = 4.13ILNSDD310 pKa = 4.59FSSAGNASGGWTLSAGTGFSSTLSDD335 pKa = 3.13GGEE338 pKa = 4.0TIEE341 pKa = 5.3FYY343 pKa = 10.54IRR345 pKa = 11.84YY346 pKa = 9.32YY347 pKa = 9.81PXNFDD352 pKa = 3.96NLMQVALTSEE362 pKa = 4.67FNMSNVPEE370 pKa = 4.26EE371 pKa = 4.13KK372 pKa = 10.18QGDD375 pKa = 4.34VIVPDD380 pKa = 3.79LTKK383 pKa = 10.06LTAYY387 pKa = 9.84KK388 pKa = 10.49DD389 pKa = 2.8IGVIVKK395 pKa = 9.98EE396 pKa = 3.96VDD398 pKa = 3.41SVSDD402 pKa = 3.33YY403 pKa = 11.0TYY405 pKa = 10.4TGKK408 pKa = 10.15HH409 pKa = 3.99STGFVLDD416 pKa = 3.61TTPNEE421 pKa = 4.27NIVYY425 pKa = 9.12TSKK428 pKa = 11.08LDD430 pKa = 3.56STVTGGLSNDD440 pKa = 3.18TMYY443 pKa = 11.48GSIGNDD449 pKa = 3.0TLEE452 pKa = 4.48GSNGNDD458 pKa = 3.39TLSGGAGVNTLIGGSGIDD476 pKa = 3.2TADD479 pKa = 3.18YY480 pKa = 11.46SFVTKK485 pKa = 10.88YY486 pKa = 11.1SDD488 pKa = 4.42DD489 pKa = 3.72FLFANDD495 pKa = 3.52GHH497 pKa = 6.94IDD499 pKa = 3.68PVTGVAYY506 pKa = 10.35ANDD509 pKa = 3.91SKK511 pKa = 11.64GVIVDD516 pKa = 4.25LQAGAASGKK525 pKa = 7.32TVYY528 pKa = 10.56DD529 pKa = 3.39ATSDD533 pKa = 3.66VLTGEE538 pKa = 4.3EE539 pKa = 4.11SRR541 pKa = 11.84TISDD545 pKa = 3.52HH546 pKa = 6.69LSGIEE551 pKa = 3.75YY552 pKa = 10.66VVGSKK557 pKa = 10.84YY558 pKa = 10.93DD559 pKa = 3.34DD560 pKa = 4.28TIRR563 pKa = 11.84GDD565 pKa = 3.46ANANKK570 pKa = 10.35LSGGEE575 pKa = 4.2GNDD578 pKa = 3.53TLEE581 pKa = 4.68GRR583 pKa = 11.84DD584 pKa = 3.54GKK586 pKa = 10.71DD587 pKa = 2.77WLEE590 pKa = 4.27GGAGNDD596 pKa = 3.41WLIGSTDD603 pKa = 3.78DD604 pKa = 4.19YY605 pKa = 11.13MIDD608 pKa = 4.1GGDD611 pKa = 3.63NTDD614 pKa = 3.58TADD617 pKa = 4.3FSNNSNGLIITLNDD631 pKa = 3.45SANGSLGNIGGTSATTIIKK650 pKa = 10.21NVEE653 pKa = 4.06NVAGTQYY660 pKa = 11.09KK661 pKa = 10.69DD662 pKa = 3.69SISGDD667 pKa = 3.45TADD670 pKa = 3.99NLLIGGYY677 pKa = 8.44DD678 pKa = 3.41HH679 pKa = 7.47SATAVTANDD688 pKa = 3.51DD689 pKa = 4.28TITGGSGADD698 pKa = 3.56TIVGDD703 pKa = 3.79MMIDD707 pKa = 3.54TVIATPLYY715 pKa = 10.59AGNDD719 pKa = 3.67VLGGGSDD726 pKa = 3.51NDD728 pKa = 4.3TIYY731 pKa = 11.34GDD733 pKa = 3.78SAPILSNAEE742 pKa = 3.82VEE744 pKa = 4.64VAADD748 pKa = 3.34QSVEE752 pKa = 4.22YY753 pKa = 10.36IRR755 pKa = 11.84NTDD758 pKa = 3.85DD759 pKa = 5.19DD760 pKa = 4.25STLATIYY767 pKa = 10.76GGNDD771 pKa = 3.16TLKK774 pKa = 11.09GGGGDD779 pKa = 3.93DD780 pKa = 3.89YY781 pKa = 11.87LNGGSGWDD789 pKa = 3.5TVDD792 pKa = 3.7YY793 pKa = 10.16SASAAAVIVNLSSNYY808 pKa = 10.77AMGEE812 pKa = 4.29GYY814 pKa = 10.29DD815 pKa = 3.27QLFNIEE821 pKa = 4.82NIVGSNSALGDD832 pKa = 3.76TLIGNALANTIVGSTSGADD851 pKa = 3.46TLSGMGGLDD860 pKa = 3.38TLNGLGGKK868 pKa = 9.16DD869 pKa = 3.01WADD872 pKa = 3.31YY873 pKa = 10.79SYY875 pKa = 11.96VSGTTGVNVDD885 pKa = 3.95LTAGTAYY892 pKa = 10.62VSATDD897 pKa = 3.85GDD899 pKa = 4.01TLIAIEE905 pKa = 4.11NVIGSSKK912 pKa = 11.18ADD914 pKa = 3.66TLSGGSGNSVNTLMGGLGNDD934 pKa = 2.7IFYY937 pKa = 10.54GYY939 pKa = 11.14GDD941 pKa = 4.31GDD943 pKa = 4.29LLDD946 pKa = 4.96GGSGSDD952 pKa = 3.19TADD955 pKa = 3.03YY956 pKa = 9.17MYY958 pKa = 10.8AQGRR962 pKa = 11.84IVVTLDD968 pKa = 3.22GSNSDD973 pKa = 3.01TLMNIEE979 pKa = 4.19NVYY982 pKa = 10.26GASDD986 pKa = 3.95KK987 pKa = 10.63DD988 pKa = 3.81TLIGNSDD995 pKa = 3.54ANILDD1000 pKa = 3.56GRR1002 pKa = 11.84ADD1004 pKa = 3.79TDD1006 pKa = 3.78TLSGMGGADD1015 pKa = 3.53TLIGGAGDD1023 pKa = 3.53DD1024 pKa = 4.13TLIGGAGNDD1033 pKa = 4.2TLWGGTDD1040 pKa = 3.24ATHH1043 pKa = 8.06DD1044 pKa = 3.98SGIDD1048 pKa = 3.18TADD1051 pKa = 3.4YY1052 pKa = 10.62SGSSSIYY1059 pKa = 10.32ADD1061 pKa = 3.66LANGTVNDD1069 pKa = 4.43GLGGIDD1075 pKa = 3.34TLAGIEE1081 pKa = 4.34VILGSGSNDD1090 pKa = 3.15VMIGSGNADD1099 pKa = 3.46TLIGNGGNDD1108 pKa = 3.16TFFASGGGDD1117 pKa = 3.04SYY1119 pKa = 11.78YY1120 pKa = 11.16GGVFGSSSDD1129 pKa = 3.65GNVKK1133 pKa = 10.46DD1134 pKa = 3.74RR1135 pKa = 11.84LSYY1138 pKa = 11.09ADD1140 pKa = 4.04VYY1142 pKa = 11.34HH1143 pKa = 7.34DD1144 pKa = 4.61VIDD1147 pKa = 4.27SKK1149 pKa = 10.07TVDD1152 pKa = 4.19HH1153 pKa = 6.67IVVDD1157 pKa = 5.01LSHH1160 pKa = 6.92VGTNATILDD1169 pKa = 3.76ASNAVISTDD1178 pKa = 2.83SLYY1181 pKa = 10.84AIEE1184 pKa = 5.43EE1185 pKa = 4.16IVATDD1190 pKa = 3.64GNDD1193 pKa = 3.52TLRR1196 pKa = 11.84GGAGSQQTFYY1206 pKa = 11.34GGAGDD1211 pKa = 4.54DD1212 pKa = 3.87TLSGNTDD1219 pKa = 2.74GDD1221 pKa = 4.11YY1222 pKa = 11.5LXTNLADD1229 pKa = 3.58YY1230 pKa = 7.63TAQSSNLVVNLASTSNNVYY1249 pKa = 10.65LSGSSPTSSNSDD1261 pKa = 2.93TLANINNIATGSGADD1276 pKa = 3.63TIIGNGNDD1284 pKa = 3.53NIVRR1288 pKa = 11.84AGVGADD1294 pKa = 3.62TLSGGAGDD1302 pKa = 3.63DD1303 pKa = 3.69TLYY1306 pKa = 11.52GEE1308 pKa = 5.21AGNDD1312 pKa = 3.54TLSGGEE1318 pKa = 4.39GNDD1321 pKa = 3.55TLIGGMSTSVDD1332 pKa = 3.57SGIDD1336 pKa = 3.28TADD1339 pKa = 3.47YY1340 pKa = 11.04SATSYY1345 pKa = 11.43YY1346 pKa = 10.69GVTVDD1351 pKa = 4.72LQNGNATGATIGTDD1365 pKa = 2.84KK1366 pKa = 11.15LYY1368 pKa = 11.02GIEE1371 pKa = 4.11NVLGSAQSDD1380 pKa = 3.92TLIGASGTLNTLSGLGGNDD1399 pKa = 2.99TLYY1402 pKa = 11.53GNLEE1406 pKa = 3.76GDD1408 pKa = 4.04YY1409 pKa = 10.95LDD1411 pKa = 5.23GGSGTNTADD1420 pKa = 3.6YY1421 pKa = 10.85SSEE1424 pKa = 4.02TASIVANLTTGRR1436 pKa = 11.84AYY1438 pKa = 8.58FTSSSSYY1445 pKa = 10.66DD1446 pKa = 3.37ALVSIQNVLSGSGNDD1461 pKa = 3.65TLIGLSGTLNTLNGNDD1477 pKa = 4.39GDD1479 pKa = 4.36DD1480 pKa = 3.84TLSGNLDD1487 pKa = 3.58GDD1489 pKa = 4.21SLIGGDD1495 pKa = 4.72GSDD1498 pKa = 3.38TLDD1501 pKa = 3.35YY1502 pKa = 7.98TTANAALTVDD1512 pKa = 5.08LGTQTLFKK1520 pKa = 10.38SASSSAKK1527 pKa = 10.04DD1528 pKa = 3.71YY1529 pKa = 10.89YY1530 pKa = 11.29SEE1532 pKa = 5.19AEE1534 pKa = 4.3VIVTGTYY1541 pKa = 10.33NDD1543 pKa = 3.82SFTLSTSTDD1552 pKa = 3.37TAAMTLDD1559 pKa = 3.83GGAGG1563 pKa = 3.22
Molecular weight: 159.4 kDa
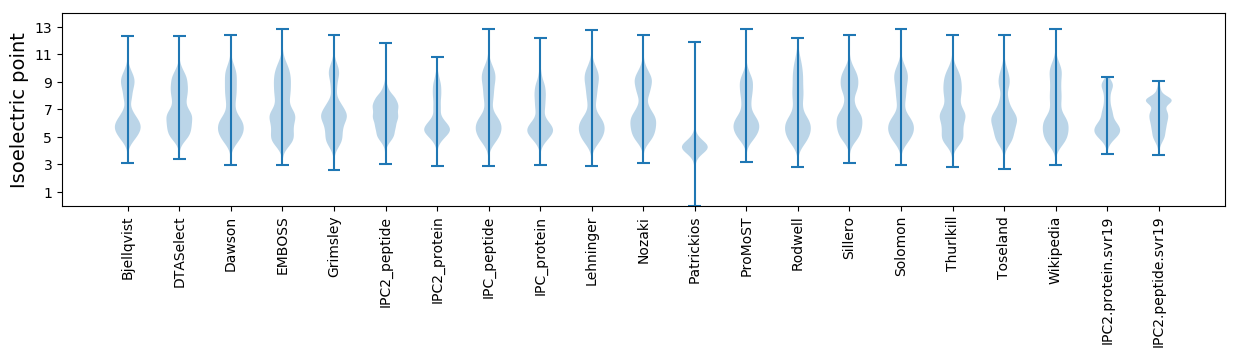
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D3W529|A0A2D3W529_9PROT Uncharacterized protein OS=Sulfurospirillum cavolei OX=366522 GN=CFH80_04675 PE=4 SV=1
MM1 pKa = 6.73TTMEE5 pKa = 4.73KK6 pKa = 9.97IKK8 pKa = 10.74FEE10 pKa = 4.5SKK12 pKa = 8.79SHH14 pKa = 5.22FRR16 pKa = 11.84AFARR20 pKa = 11.84AKK22 pKa = 10.78LEE24 pKa = 4.07TLNGCYY30 pKa = 9.93RR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.53KK34 pKa = 10.95LLQRR38 pKa = 11.84QLSSLLFHH46 pKa = 6.11MRR48 pKa = 11.84VRR50 pKa = 11.84SILLYY55 pKa = 10.68NPLAIEE61 pKa = 3.89ADD63 pKa = 3.37IRR65 pKa = 11.84SVIHH69 pKa = 4.29TCRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 10.23CKK77 pKa = 9.93VHH79 pKa = 6.19VPFMEE84 pKa = 5.25GISFKK89 pKa = 9.99MVEE92 pKa = 3.6WRR94 pKa = 11.84LPLEE98 pKa = 3.9KK99 pKa = 9.83RR100 pKa = 11.84VFNILEE106 pKa = 4.25PKK108 pKa = 10.56NSFKK112 pKa = 11.0LKK114 pKa = 10.29PKK116 pKa = 9.8IDD118 pKa = 3.82LAVVPVIGVDD128 pKa = 3.66GAFKK132 pKa = 10.63RR133 pKa = 11.84VGFGKK138 pKa = 10.6GMYY141 pKa = 10.17DD142 pKa = 4.29RR143 pKa = 11.84FFQTLAKK150 pKa = 9.83KK151 pKa = 10.08PPIVFIQRR159 pKa = 11.84EE160 pKa = 4.08LCMTQSRR167 pKa = 11.84LSEE170 pKa = 4.12EE171 pKa = 3.9HH172 pKa = 7.44DD173 pKa = 3.31IAADD177 pKa = 3.46IYY179 pKa = 8.69LTPYY183 pKa = 8.5HH184 pKa = 6.41TIIKK188 pKa = 9.93RR189 pKa = 11.84GEE191 pKa = 3.76NGPRR195 pKa = 11.84VVSRR199 pKa = 11.84RR200 pKa = 11.84WSRR203 pKa = 11.84ISQRR207 pKa = 11.84RR208 pKa = 11.84CRR210 pKa = 11.84IFHH213 pKa = 5.73RR214 pKa = 11.84QEE216 pKa = 4.06DD217 pKa = 4.29GSRR220 pKa = 11.84KK221 pKa = 9.5LL222 pKa = 3.54
MM1 pKa = 6.73TTMEE5 pKa = 4.73KK6 pKa = 9.97IKK8 pKa = 10.74FEE10 pKa = 4.5SKK12 pKa = 8.79SHH14 pKa = 5.22FRR16 pKa = 11.84AFARR20 pKa = 11.84AKK22 pKa = 10.78LEE24 pKa = 4.07TLNGCYY30 pKa = 9.93RR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.53KK34 pKa = 10.95LLQRR38 pKa = 11.84QLSSLLFHH46 pKa = 6.11MRR48 pKa = 11.84VRR50 pKa = 11.84SILLYY55 pKa = 10.68NPLAIEE61 pKa = 3.89ADD63 pKa = 3.37IRR65 pKa = 11.84SVIHH69 pKa = 4.29TCRR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 10.23CKK77 pKa = 9.93VHH79 pKa = 6.19VPFMEE84 pKa = 5.25GISFKK89 pKa = 9.99MVEE92 pKa = 3.6WRR94 pKa = 11.84LPLEE98 pKa = 3.9KK99 pKa = 9.83RR100 pKa = 11.84VFNILEE106 pKa = 4.25PKK108 pKa = 10.56NSFKK112 pKa = 11.0LKK114 pKa = 10.29PKK116 pKa = 9.8IDD118 pKa = 3.82LAVVPVIGVDD128 pKa = 3.66GAFKK132 pKa = 10.63RR133 pKa = 11.84VGFGKK138 pKa = 10.6GMYY141 pKa = 10.17DD142 pKa = 4.29RR143 pKa = 11.84FFQTLAKK150 pKa = 9.83KK151 pKa = 10.08PPIVFIQRR159 pKa = 11.84EE160 pKa = 4.08LCMTQSRR167 pKa = 11.84LSEE170 pKa = 4.12EE171 pKa = 3.9HH172 pKa = 7.44DD173 pKa = 3.31IAADD177 pKa = 3.46IYY179 pKa = 8.69LTPYY183 pKa = 8.5HH184 pKa = 6.41TIIKK188 pKa = 9.93RR189 pKa = 11.84GEE191 pKa = 3.76NGPRR195 pKa = 11.84VVSRR199 pKa = 11.84RR200 pKa = 11.84WSRR203 pKa = 11.84ISQRR207 pKa = 11.84RR208 pKa = 11.84CRR210 pKa = 11.84IFHH213 pKa = 5.73RR214 pKa = 11.84QEE216 pKa = 4.06DD217 pKa = 4.29GSRR220 pKa = 11.84KK221 pKa = 9.5LL222 pKa = 3.54
Molecular weight: 26.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
538432 |
26 |
1563 |
287.3 |
32.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.255 ± 0.06 | 1.158 ± 0.024 |
5.115 ± 0.046 | 6.873 ± 0.066 |
5.019 ± 0.051 | 6.202 ± 0.061 |
2.326 ± 0.03 | 7.38 ± 0.048 |
6.989 ± 0.062 | 10.412 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.679 ± 0.023 | 4.111 ± 0.037 |
3.259 ± 0.035 | 3.288 ± 0.031 |
4.02 ± 0.037 | 6.432 ± 0.045 |
5.284 ± 0.041 | 6.656 ± 0.046 |
0.838 ± 0.018 | 3.687 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
