
Macaca mulatta feces associated virus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus macas2
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
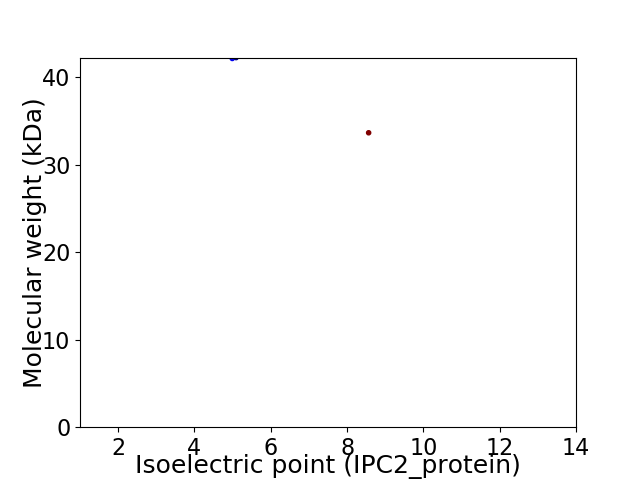
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
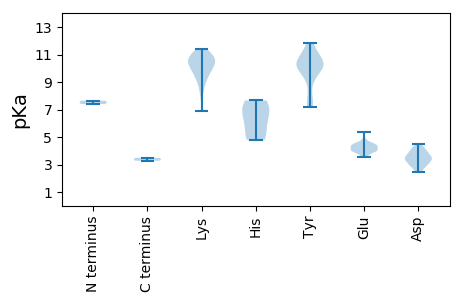
Protein with the lowest isoelectric point:
>tr|A0A1W5Q0A5|A0A1W5Q0A5_9VIRU Rep OS=Macaca mulatta feces associated virus 4 OX=2499226 PE=4 SV=1
MM1 pKa = 7.64LPEE4 pKa = 4.53GLTAEE9 pKa = 4.39GTIEE13 pKa = 3.98GDD15 pKa = 3.03RR16 pKa = 11.84VVSVRR21 pKa = 11.84VSEE24 pKa = 4.47TYY26 pKa = 10.7DD27 pKa = 3.52LSTQVGKK34 pKa = 9.88MGIVGIHH41 pKa = 5.32TPIGTLVEE49 pKa = 4.4RR50 pKa = 11.84MWSGLVRR57 pKa = 11.84QYY59 pKa = 11.8GKK61 pKa = 10.68FRR63 pKa = 11.84FKK65 pKa = 11.02SCDD68 pKa = 3.09VAMACASMLPADD80 pKa = 4.26PLQIGVEE87 pKa = 4.06AGAIAPQDD95 pKa = 3.54MFNPILYY102 pKa = 9.84KK103 pKa = 10.59AVSNDD108 pKa = 3.03TMNTFLQYY116 pKa = 10.4IQGQAIFNTGSTSGKK131 pKa = 8.76IVNFGSVIGVNDD143 pKa = 4.11SDD145 pKa = 4.19FKK147 pKa = 11.03TPGDD151 pKa = 3.93APTSIDD157 pKa = 3.6QFQMYY162 pKa = 10.03YY163 pKa = 11.12GLLSDD168 pKa = 3.82PSGWRR173 pKa = 11.84KK174 pKa = 9.98AMPQSGLVMRR184 pKa = 11.84GLRR187 pKa = 11.84PLVYY191 pKa = 10.35NVVSSYY197 pKa = 11.31GLNEE201 pKa = 3.73ASGAGVTGAPKK212 pKa = 10.53NDD214 pKa = 2.85IKK216 pKa = 11.37SNQFIGDD223 pKa = 3.55TRR225 pKa = 11.84YY226 pKa = 7.41TAAGARR232 pKa = 11.84AATNGSDD239 pKa = 3.32AQLPDD244 pKa = 3.84VVSGGQNVYY253 pKa = 10.51QDD255 pKa = 3.65NPVFGSFKK263 pKa = 10.54GRR265 pKa = 11.84SQPMPWINTKK275 pKa = 9.73FWPSAVGNSDD285 pKa = 2.46GSMITEE291 pKa = 4.03TEE293 pKa = 4.22STNKK297 pKa = 10.36DD298 pKa = 2.69ATLQSNVGRR307 pKa = 11.84VPPAYY312 pKa = 10.09VGLIVLPPAKK322 pKa = 10.23LNQLYY327 pKa = 10.29YY328 pKa = 10.16RR329 pKa = 11.84IKK331 pKa = 9.09VTWTIEE337 pKa = 3.97FSEE340 pKa = 4.45LQSNLAVSNWQGIAKK355 pKa = 10.11FGSLSYY361 pKa = 9.92ATDD364 pKa = 3.31YY365 pKa = 11.21AAQSKK370 pKa = 10.99SMDD373 pKa = 4.26SIAGMVDD380 pKa = 2.87TSDD383 pKa = 4.03ADD385 pKa = 3.47VEE387 pKa = 4.59KK388 pKa = 10.81IMEE391 pKa = 4.41GAA393 pKa = 3.49
MM1 pKa = 7.64LPEE4 pKa = 4.53GLTAEE9 pKa = 4.39GTIEE13 pKa = 3.98GDD15 pKa = 3.03RR16 pKa = 11.84VVSVRR21 pKa = 11.84VSEE24 pKa = 4.47TYY26 pKa = 10.7DD27 pKa = 3.52LSTQVGKK34 pKa = 9.88MGIVGIHH41 pKa = 5.32TPIGTLVEE49 pKa = 4.4RR50 pKa = 11.84MWSGLVRR57 pKa = 11.84QYY59 pKa = 11.8GKK61 pKa = 10.68FRR63 pKa = 11.84FKK65 pKa = 11.02SCDD68 pKa = 3.09VAMACASMLPADD80 pKa = 4.26PLQIGVEE87 pKa = 4.06AGAIAPQDD95 pKa = 3.54MFNPILYY102 pKa = 9.84KK103 pKa = 10.59AVSNDD108 pKa = 3.03TMNTFLQYY116 pKa = 10.4IQGQAIFNTGSTSGKK131 pKa = 8.76IVNFGSVIGVNDD143 pKa = 4.11SDD145 pKa = 4.19FKK147 pKa = 11.03TPGDD151 pKa = 3.93APTSIDD157 pKa = 3.6QFQMYY162 pKa = 10.03YY163 pKa = 11.12GLLSDD168 pKa = 3.82PSGWRR173 pKa = 11.84KK174 pKa = 9.98AMPQSGLVMRR184 pKa = 11.84GLRR187 pKa = 11.84PLVYY191 pKa = 10.35NVVSSYY197 pKa = 11.31GLNEE201 pKa = 3.73ASGAGVTGAPKK212 pKa = 10.53NDD214 pKa = 2.85IKK216 pKa = 11.37SNQFIGDD223 pKa = 3.55TRR225 pKa = 11.84YY226 pKa = 7.41TAAGARR232 pKa = 11.84AATNGSDD239 pKa = 3.32AQLPDD244 pKa = 3.84VVSGGQNVYY253 pKa = 10.51QDD255 pKa = 3.65NPVFGSFKK263 pKa = 10.54GRR265 pKa = 11.84SQPMPWINTKK275 pKa = 9.73FWPSAVGNSDD285 pKa = 2.46GSMITEE291 pKa = 4.03TEE293 pKa = 4.22STNKK297 pKa = 10.36DD298 pKa = 2.69ATLQSNVGRR307 pKa = 11.84VPPAYY312 pKa = 10.09VGLIVLPPAKK322 pKa = 10.23LNQLYY327 pKa = 10.29YY328 pKa = 10.16RR329 pKa = 11.84IKK331 pKa = 9.09VTWTIEE337 pKa = 3.97FSEE340 pKa = 4.45LQSNLAVSNWQGIAKK355 pKa = 10.11FGSLSYY361 pKa = 9.92ATDD364 pKa = 3.31YY365 pKa = 11.21AAQSKK370 pKa = 10.99SMDD373 pKa = 4.26SIAGMVDD380 pKa = 2.87TSDD383 pKa = 4.03ADD385 pKa = 3.47VEE387 pKa = 4.59KK388 pKa = 10.81IMEE391 pKa = 4.41GAA393 pKa = 3.49
Molecular weight: 42.13 kDa
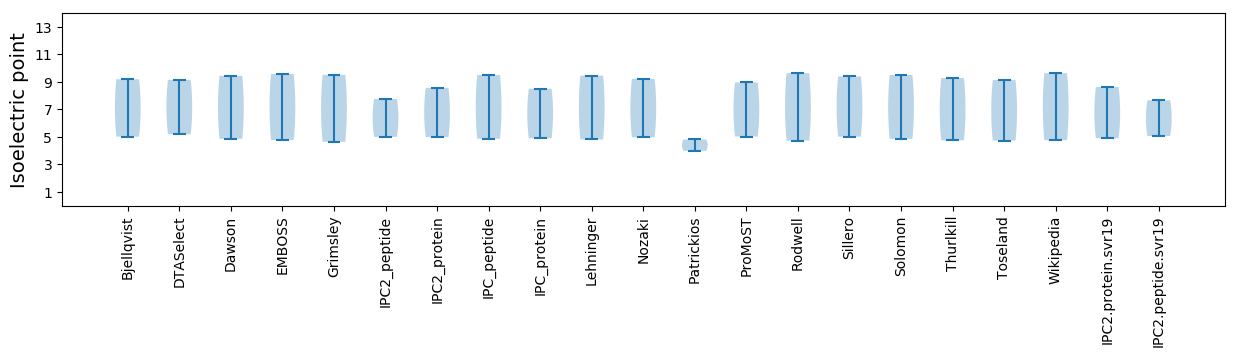
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5Q0A5|A0A1W5Q0A5_9VIRU Rep OS=Macaca mulatta feces associated virus 4 OX=2499226 PE=4 SV=1
MM1 pKa = 7.4RR2 pKa = 11.84RR3 pKa = 11.84IRR5 pKa = 11.84EE6 pKa = 4.39TMGKK10 pKa = 8.04TYY12 pKa = 10.37MLTIPRR18 pKa = 11.84NDD20 pKa = 3.42TYY22 pKa = 11.52RR23 pKa = 11.84EE24 pKa = 4.14GNKK27 pKa = 9.27IFAWFRR33 pKa = 11.84KK34 pKa = 9.35NDD36 pKa = 2.9IHH38 pKa = 7.66KK39 pKa = 9.66WVLGAEE45 pKa = 4.32KK46 pKa = 10.65GSGGYY51 pKa = 7.21EE52 pKa = 3.54HH53 pKa = 6.86WQMRR57 pKa = 11.84VQCRR61 pKa = 11.84FGFEE65 pKa = 3.92EE66 pKa = 4.85LKK68 pKa = 11.12VLFPTAHH75 pKa = 6.85IEE77 pKa = 4.06EE78 pKa = 5.34CSDD81 pKa = 2.43KK82 pKa = 9.17WTYY85 pKa = 7.48EE86 pKa = 3.98AKK88 pKa = 10.63EE89 pKa = 3.96GVYY92 pKa = 9.06WRR94 pKa = 11.84SNDD97 pKa = 2.95RR98 pKa = 11.84RR99 pKa = 11.84EE100 pKa = 4.38NIQQRR105 pKa = 11.84FGKK108 pKa = 9.99LRR110 pKa = 11.84YY111 pKa = 7.83AQKK114 pKa = 10.56RR115 pKa = 11.84VIEE118 pKa = 4.38RR119 pKa = 11.84ADD121 pKa = 3.38GTNDD125 pKa = 3.28RR126 pKa = 11.84EE127 pKa = 4.38VVVWYY132 pKa = 9.72DD133 pKa = 3.37QEE135 pKa = 4.44GKK137 pKa = 10.31AGKK140 pKa = 9.48SWLTGAMWEE149 pKa = 4.15RR150 pKa = 11.84GLAYY154 pKa = 10.13FSVADD159 pKa = 3.9TSGKK163 pKa = 8.42TIVMDD168 pKa = 4.01IASEE172 pKa = 4.03YY173 pKa = 10.46LKK175 pKa = 11.06NGWRR179 pKa = 11.84PYY181 pKa = 10.32IIIDD185 pKa = 3.45IPRR188 pKa = 11.84AGKK191 pKa = 6.86WTPEE195 pKa = 3.79LYY197 pKa = 10.39EE198 pKa = 4.67AIEE201 pKa = 4.45RR202 pKa = 11.84IKK204 pKa = 11.02DD205 pKa = 3.65GLIKK209 pKa = 10.41DD210 pKa = 3.72PRR212 pKa = 11.84YY213 pKa = 10.31SSEE216 pKa = 3.54AVNIHH221 pKa = 4.8GVKK224 pKa = 10.15VIVMTNTMPKK234 pKa = 9.79LDD236 pKa = 4.46KK237 pKa = 11.01LSADD241 pKa = 2.96RR242 pKa = 11.84WKK244 pKa = 10.6IITQQEE250 pKa = 4.09LTLEE254 pKa = 4.13EE255 pKa = 3.89TAIEE259 pKa = 4.08QFALTGKK266 pKa = 10.13GGYY269 pKa = 9.12RR270 pKa = 11.84PPLTPPTWDD279 pKa = 3.28SRR281 pKa = 11.84DD282 pKa = 3.3VRR284 pKa = 11.84GPLSS288 pKa = 3.28
MM1 pKa = 7.4RR2 pKa = 11.84RR3 pKa = 11.84IRR5 pKa = 11.84EE6 pKa = 4.39TMGKK10 pKa = 8.04TYY12 pKa = 10.37MLTIPRR18 pKa = 11.84NDD20 pKa = 3.42TYY22 pKa = 11.52RR23 pKa = 11.84EE24 pKa = 4.14GNKK27 pKa = 9.27IFAWFRR33 pKa = 11.84KK34 pKa = 9.35NDD36 pKa = 2.9IHH38 pKa = 7.66KK39 pKa = 9.66WVLGAEE45 pKa = 4.32KK46 pKa = 10.65GSGGYY51 pKa = 7.21EE52 pKa = 3.54HH53 pKa = 6.86WQMRR57 pKa = 11.84VQCRR61 pKa = 11.84FGFEE65 pKa = 3.92EE66 pKa = 4.85LKK68 pKa = 11.12VLFPTAHH75 pKa = 6.85IEE77 pKa = 4.06EE78 pKa = 5.34CSDD81 pKa = 2.43KK82 pKa = 9.17WTYY85 pKa = 7.48EE86 pKa = 3.98AKK88 pKa = 10.63EE89 pKa = 3.96GVYY92 pKa = 9.06WRR94 pKa = 11.84SNDD97 pKa = 2.95RR98 pKa = 11.84RR99 pKa = 11.84EE100 pKa = 4.38NIQQRR105 pKa = 11.84FGKK108 pKa = 9.99LRR110 pKa = 11.84YY111 pKa = 7.83AQKK114 pKa = 10.56RR115 pKa = 11.84VIEE118 pKa = 4.38RR119 pKa = 11.84ADD121 pKa = 3.38GTNDD125 pKa = 3.28RR126 pKa = 11.84EE127 pKa = 4.38VVVWYY132 pKa = 9.72DD133 pKa = 3.37QEE135 pKa = 4.44GKK137 pKa = 10.31AGKK140 pKa = 9.48SWLTGAMWEE149 pKa = 4.15RR150 pKa = 11.84GLAYY154 pKa = 10.13FSVADD159 pKa = 3.9TSGKK163 pKa = 8.42TIVMDD168 pKa = 4.01IASEE172 pKa = 4.03YY173 pKa = 10.46LKK175 pKa = 11.06NGWRR179 pKa = 11.84PYY181 pKa = 10.32IIIDD185 pKa = 3.45IPRR188 pKa = 11.84AGKK191 pKa = 6.86WTPEE195 pKa = 3.79LYY197 pKa = 10.39EE198 pKa = 4.67AIEE201 pKa = 4.45RR202 pKa = 11.84IKK204 pKa = 11.02DD205 pKa = 3.65GLIKK209 pKa = 10.41DD210 pKa = 3.72PRR212 pKa = 11.84YY213 pKa = 10.31SSEE216 pKa = 3.54AVNIHH221 pKa = 4.8GVKK224 pKa = 10.15VIVMTNTMPKK234 pKa = 9.79LDD236 pKa = 4.46KK237 pKa = 11.01LSADD241 pKa = 2.96RR242 pKa = 11.84WKK244 pKa = 10.6IITQQEE250 pKa = 4.09LTLEE254 pKa = 4.13EE255 pKa = 3.89TAIEE259 pKa = 4.08QFALTGKK266 pKa = 10.13GGYY269 pKa = 9.12RR270 pKa = 11.84PPLTPPTWDD279 pKa = 3.28SRR281 pKa = 11.84DD282 pKa = 3.3VRR284 pKa = 11.84GPLSS288 pKa = 3.28
Molecular weight: 33.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
681 |
288 |
393 |
340.5 |
37.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.342 ± 0.925 | 0.587 ± 0.069 |
5.727 ± 0.11 | 5.433 ± 1.864 |
3.231 ± 0.291 | 9.251 ± 0.813 |
0.734 ± 0.421 | 6.314 ± 0.628 |
5.727 ± 1.229 | 6.021 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.377 ± 0.385 | 4.258 ± 0.728 |
4.846 ± 0.436 | 4.258 ± 0.728 |
5.58 ± 1.992 | 7.195 ± 1.946 |
6.461 ± 0.087 | 6.902 ± 1.088 |
2.643 ± 0.979 | 4.112 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
