
Hubei tombus-like virus 25
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
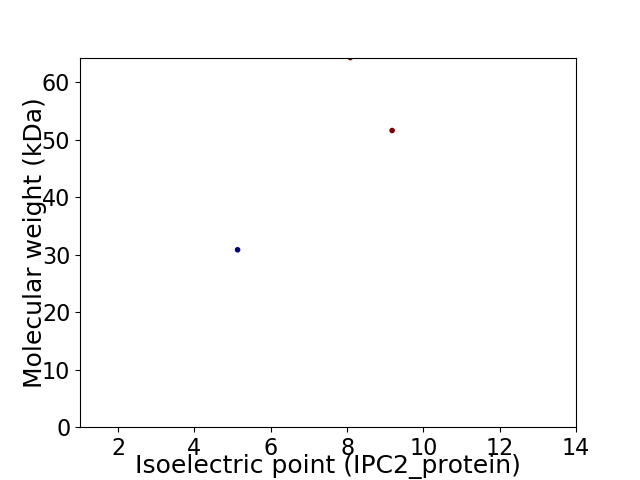
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
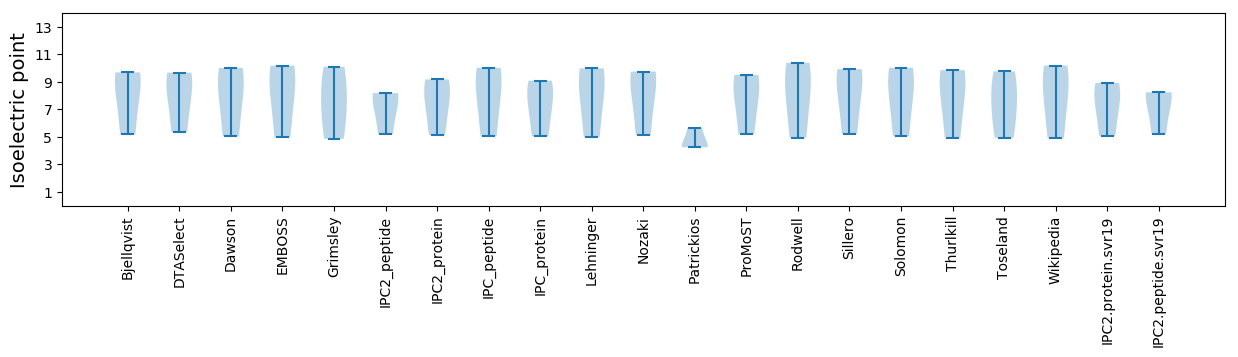
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGV5|A0A1L3KGV5_9VIRU Capsid protein OS=Hubei tombus-like virus 25 OX=1923272 PE=3 SV=1
MM1 pKa = 7.77FDD3 pKa = 3.7VLVDD7 pKa = 3.54NCAVDD12 pKa = 4.32EE13 pKa = 4.9DD14 pKa = 5.06PNDD17 pKa = 3.64VMNLPGVAVHH27 pKa = 6.58MDD29 pKa = 3.33AEE31 pKa = 4.22NAANYY36 pKa = 9.01INSIIEE42 pKa = 3.98PGAKK46 pKa = 9.61RR47 pKa = 11.84MGKK50 pKa = 9.72NKK52 pKa = 9.87IEE54 pKa = 4.08GLNAVSVGKK63 pKa = 10.46RR64 pKa = 11.84KK65 pKa = 9.54EE66 pKa = 4.04VVDD69 pKa = 4.06AVCGIFEE76 pKa = 5.3DD77 pKa = 4.11SANDD81 pKa = 2.98IDD83 pKa = 5.71FGIKK87 pKa = 10.54LMTSGAKK94 pKa = 10.01ARR96 pKa = 11.84IAEE99 pKa = 4.24AVNVAMDD106 pKa = 3.92EE107 pKa = 4.32AGGPAMVPVVKK118 pKa = 10.09EE119 pKa = 4.22VYY121 pKa = 8.11TQASVGCEE129 pKa = 3.92TGVGTDD135 pKa = 3.05DD136 pKa = 3.25VTRR139 pKa = 11.84PRR141 pKa = 11.84RR142 pKa = 11.84AFRR145 pKa = 11.84VQKK148 pKa = 10.56RR149 pKa = 11.84RR150 pKa = 11.84DD151 pKa = 3.32FRR153 pKa = 11.84SYY155 pKa = 11.16RR156 pKa = 11.84NAGCIEE162 pKa = 4.09ADD164 pKa = 3.47EE165 pKa = 4.29TLTYY169 pKa = 10.62YY170 pKa = 10.26LKK172 pKa = 10.63CKK174 pKa = 10.56YY175 pKa = 10.03FMKK178 pKa = 10.39PRR180 pKa = 11.84DD181 pKa = 3.96SGTLHH186 pKa = 6.44QMVSDD191 pKa = 4.24ARR193 pKa = 11.84VQMMKK198 pKa = 10.2SGHH201 pKa = 5.39TLDD204 pKa = 4.93SEE206 pKa = 4.42VDD208 pKa = 3.51YY209 pKa = 11.64EE210 pKa = 4.3MMTRR214 pKa = 11.84AVMAAFLVDD223 pKa = 3.37AAEE226 pKa = 4.1LEE228 pKa = 4.19FRR230 pKa = 11.84QRR232 pKa = 11.84LKK234 pKa = 11.23NYY236 pKa = 9.81DD237 pKa = 3.57AFDD240 pKa = 3.88AATKK244 pKa = 8.82ITEE247 pKa = 4.22AANGNLGRR255 pKa = 11.84IDD257 pKa = 4.32LLGARR262 pKa = 11.84AGQGSILGKK271 pKa = 9.25MLPSLDD277 pKa = 4.22LDD279 pKa = 4.0TPKK282 pKa = 10.75VV283 pKa = 3.44
MM1 pKa = 7.77FDD3 pKa = 3.7VLVDD7 pKa = 3.54NCAVDD12 pKa = 4.32EE13 pKa = 4.9DD14 pKa = 5.06PNDD17 pKa = 3.64VMNLPGVAVHH27 pKa = 6.58MDD29 pKa = 3.33AEE31 pKa = 4.22NAANYY36 pKa = 9.01INSIIEE42 pKa = 3.98PGAKK46 pKa = 9.61RR47 pKa = 11.84MGKK50 pKa = 9.72NKK52 pKa = 9.87IEE54 pKa = 4.08GLNAVSVGKK63 pKa = 10.46RR64 pKa = 11.84KK65 pKa = 9.54EE66 pKa = 4.04VVDD69 pKa = 4.06AVCGIFEE76 pKa = 5.3DD77 pKa = 4.11SANDD81 pKa = 2.98IDD83 pKa = 5.71FGIKK87 pKa = 10.54LMTSGAKK94 pKa = 10.01ARR96 pKa = 11.84IAEE99 pKa = 4.24AVNVAMDD106 pKa = 3.92EE107 pKa = 4.32AGGPAMVPVVKK118 pKa = 10.09EE119 pKa = 4.22VYY121 pKa = 8.11TQASVGCEE129 pKa = 3.92TGVGTDD135 pKa = 3.05DD136 pKa = 3.25VTRR139 pKa = 11.84PRR141 pKa = 11.84RR142 pKa = 11.84AFRR145 pKa = 11.84VQKK148 pKa = 10.56RR149 pKa = 11.84RR150 pKa = 11.84DD151 pKa = 3.32FRR153 pKa = 11.84SYY155 pKa = 11.16RR156 pKa = 11.84NAGCIEE162 pKa = 4.09ADD164 pKa = 3.47EE165 pKa = 4.29TLTYY169 pKa = 10.62YY170 pKa = 10.26LKK172 pKa = 10.63CKK174 pKa = 10.56YY175 pKa = 10.03FMKK178 pKa = 10.39PRR180 pKa = 11.84DD181 pKa = 3.96SGTLHH186 pKa = 6.44QMVSDD191 pKa = 4.24ARR193 pKa = 11.84VQMMKK198 pKa = 10.2SGHH201 pKa = 5.39TLDD204 pKa = 4.93SEE206 pKa = 4.42VDD208 pKa = 3.51YY209 pKa = 11.64EE210 pKa = 4.3MMTRR214 pKa = 11.84AVMAAFLVDD223 pKa = 3.37AAEE226 pKa = 4.1LEE228 pKa = 4.19FRR230 pKa = 11.84QRR232 pKa = 11.84LKK234 pKa = 11.23NYY236 pKa = 9.81DD237 pKa = 3.57AFDD240 pKa = 3.88AATKK244 pKa = 8.82ITEE247 pKa = 4.22AANGNLGRR255 pKa = 11.84IDD257 pKa = 4.32LLGARR262 pKa = 11.84AGQGSILGKK271 pKa = 9.25MLPSLDD277 pKa = 4.22LDD279 pKa = 4.0TPKK282 pKa = 10.75VV283 pKa = 3.44
Molecular weight: 30.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGV5|A0A1L3KGV5_9VIRU Capsid protein OS=Hubei tombus-like virus 25 OX=1923272 PE=3 SV=1
MM1 pKa = 7.38NVTIQQKK8 pKa = 10.4KK9 pKa = 10.54GSTSKK14 pKa = 10.8NKK16 pKa = 9.29QRR18 pKa = 11.84SKK20 pKa = 11.0KK21 pKa = 9.62KK22 pKa = 10.28RR23 pKa = 11.84NNKK26 pKa = 7.25TSRR29 pKa = 11.84NPLSAKK35 pKa = 9.85ASKK38 pKa = 8.94GTAKK42 pKa = 10.82ANTAPVSINVTTKK55 pKa = 10.66KK56 pKa = 10.41GIPAIKK62 pKa = 8.91STAGGITVTHH72 pKa = 6.76KK73 pKa = 10.8EE74 pKa = 4.14YY75 pKa = 10.57IGDD78 pKa = 3.8FTSNGASFALNNYY91 pKa = 8.05NVNPGLSGTFPWLSAIANRR110 pKa = 11.84YY111 pKa = 7.82EE112 pKa = 4.18SYY114 pKa = 10.94LIDD117 pKa = 4.53DD118 pKa = 3.86LHH120 pKa = 7.48FIYY123 pKa = 10.67EE124 pKa = 4.66PICPTTTPGSILMAMDD140 pKa = 3.95YY141 pKa = 10.8DD142 pKa = 4.35AADD145 pKa = 3.6SAPSNKK151 pKa = 7.28VTIMAYY157 pKa = 9.8SSAVRR162 pKa = 11.84TSPWNRR168 pKa = 11.84TVFAARR174 pKa = 11.84RR175 pKa = 11.84SDD177 pKa = 3.23LHH179 pKa = 7.32KK180 pKa = 10.8FGVQRR185 pKa = 11.84YY186 pKa = 6.83VRR188 pKa = 11.84STVPPTNTDD197 pKa = 2.57VKK199 pKa = 10.02TYY201 pKa = 10.94DD202 pKa = 3.09VGNFFLASQGTPAGPTPLGEE222 pKa = 4.7LYY224 pKa = 10.73VSYY227 pKa = 9.78TIRR230 pKa = 11.84FLTPQVGTGILAPSTVMAAEE250 pKa = 4.16SQSTILRR257 pKa = 11.84IPAGVAAVNLSGEE270 pKa = 4.24YY271 pKa = 9.71RR272 pKa = 11.84GSGVNPLAWLEE283 pKa = 3.95PLAATTPDD291 pKa = 3.69PVLLLNLNNANNMLLSFRR309 pKa = 11.84AEE311 pKa = 4.15NGWGGSAAGWNPLAFFQNMRR331 pKa = 11.84MGTSVTDD338 pKa = 3.5IGVSWAVKK346 pKa = 9.56QLAFGFAEE354 pKa = 4.26KK355 pKa = 10.53NDD357 pKa = 4.17PNSLFQTYY365 pKa = 10.18LIQPTMDD372 pKa = 4.29ALSKK376 pKa = 9.92SQGGIFPLILKK387 pKa = 10.24RR388 pKa = 11.84PATGTYY394 pKa = 9.69QMAVALMPVAANYY407 pKa = 10.08SGNIPQITQMITDD420 pKa = 3.71WAFPQLYY427 pKa = 9.66IPTFNASLLTKK438 pKa = 10.42AGMRR442 pKa = 11.84ASFTDD447 pKa = 3.43TLNTIHH453 pKa = 7.43RR454 pKa = 11.84ADD456 pKa = 3.99PQDD459 pKa = 3.88TVSMIDD465 pKa = 3.61DD466 pKa = 4.86LEE468 pKa = 4.62TKK470 pKa = 10.49VKK472 pKa = 9.98TKK474 pKa = 10.81LKK476 pKa = 9.06WW477 pKa = 3.16
MM1 pKa = 7.38NVTIQQKK8 pKa = 10.4KK9 pKa = 10.54GSTSKK14 pKa = 10.8NKK16 pKa = 9.29QRR18 pKa = 11.84SKK20 pKa = 11.0KK21 pKa = 9.62KK22 pKa = 10.28RR23 pKa = 11.84NNKK26 pKa = 7.25TSRR29 pKa = 11.84NPLSAKK35 pKa = 9.85ASKK38 pKa = 8.94GTAKK42 pKa = 10.82ANTAPVSINVTTKK55 pKa = 10.66KK56 pKa = 10.41GIPAIKK62 pKa = 8.91STAGGITVTHH72 pKa = 6.76KK73 pKa = 10.8EE74 pKa = 4.14YY75 pKa = 10.57IGDD78 pKa = 3.8FTSNGASFALNNYY91 pKa = 8.05NVNPGLSGTFPWLSAIANRR110 pKa = 11.84YY111 pKa = 7.82EE112 pKa = 4.18SYY114 pKa = 10.94LIDD117 pKa = 4.53DD118 pKa = 3.86LHH120 pKa = 7.48FIYY123 pKa = 10.67EE124 pKa = 4.66PICPTTTPGSILMAMDD140 pKa = 3.95YY141 pKa = 10.8DD142 pKa = 4.35AADD145 pKa = 3.6SAPSNKK151 pKa = 7.28VTIMAYY157 pKa = 9.8SSAVRR162 pKa = 11.84TSPWNRR168 pKa = 11.84TVFAARR174 pKa = 11.84RR175 pKa = 11.84SDD177 pKa = 3.23LHH179 pKa = 7.32KK180 pKa = 10.8FGVQRR185 pKa = 11.84YY186 pKa = 6.83VRR188 pKa = 11.84STVPPTNTDD197 pKa = 2.57VKK199 pKa = 10.02TYY201 pKa = 10.94DD202 pKa = 3.09VGNFFLASQGTPAGPTPLGEE222 pKa = 4.7LYY224 pKa = 10.73VSYY227 pKa = 9.78TIRR230 pKa = 11.84FLTPQVGTGILAPSTVMAAEE250 pKa = 4.16SQSTILRR257 pKa = 11.84IPAGVAAVNLSGEE270 pKa = 4.24YY271 pKa = 9.71RR272 pKa = 11.84GSGVNPLAWLEE283 pKa = 3.95PLAATTPDD291 pKa = 3.69PVLLLNLNNANNMLLSFRR309 pKa = 11.84AEE311 pKa = 4.15NGWGGSAAGWNPLAFFQNMRR331 pKa = 11.84MGTSVTDD338 pKa = 3.5IGVSWAVKK346 pKa = 9.56QLAFGFAEE354 pKa = 4.26KK355 pKa = 10.53NDD357 pKa = 4.17PNSLFQTYY365 pKa = 10.18LIQPTMDD372 pKa = 4.29ALSKK376 pKa = 9.92SQGGIFPLILKK387 pKa = 10.24RR388 pKa = 11.84PATGTYY394 pKa = 9.69QMAVALMPVAANYY407 pKa = 10.08SGNIPQITQMITDD420 pKa = 3.71WAFPQLYY427 pKa = 9.66IPTFNASLLTKK438 pKa = 10.42AGMRR442 pKa = 11.84ASFTDD447 pKa = 3.43TLNTIHH453 pKa = 7.43RR454 pKa = 11.84ADD456 pKa = 3.99PQDD459 pKa = 3.88TVSMIDD465 pKa = 3.61DD466 pKa = 4.86LEE468 pKa = 4.62TKK470 pKa = 10.49VKK472 pKa = 9.98TKK474 pKa = 10.81LKK476 pKa = 9.06WW477 pKa = 3.16
Molecular weight: 51.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1318 |
283 |
558 |
439.3 |
48.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.877 ± 1.341 | 1.517 ± 0.561 |
5.539 ± 0.879 | 4.628 ± 0.992 |
3.718 ± 0.144 | 6.525 ± 0.515 |
1.366 ± 0.306 | 5.615 ± 0.479 |
5.918 ± 0.106 | 8.118 ± 0.595 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.338 ± 0.539 | 5.463 ± 0.595 |
4.704 ± 0.769 | 3.414 ± 0.375 |
5.842 ± 0.871 | 6.98 ± 0.725 |
6.904 ± 1.131 | 6.146 ± 0.966 |
1.442 ± 0.405 | 3.945 ± 0.551 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
