
Aythya fuligula (Tufted duck) (Anas fuligula)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria; Dinosauria; Saurischia;
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
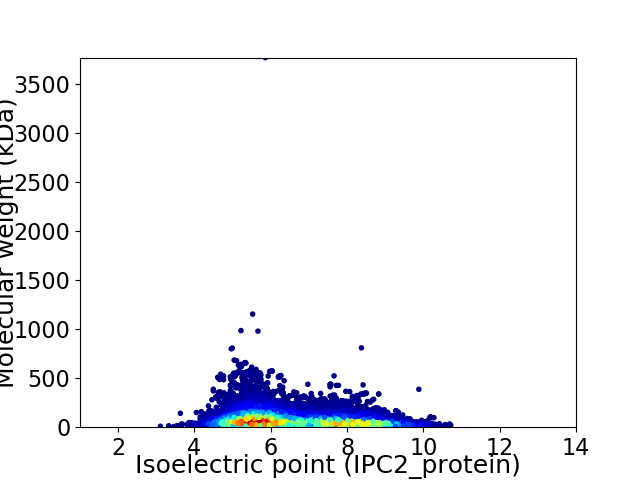
Virtual 2D-PAGE plot for 24034 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A0A6J3EPL9|A0A6J3EPL9_AYTFU glucose 1 6-bisphosphate synthase OS=Aythya fuligula OX=219594 GN=PGM2L1 PE=3 SV=1
MM1 pKa = 7.4TPNPSTTTTPDD12 pKa = 2.66ISTSTLVPSASTLTTPEE29 pKa = 3.57SSYY32 pKa = 9.68PTTPDD37 pKa = 2.87TSATTIPDD45 pKa = 3.56TSDD48 pKa = 2.73TTIPDD53 pKa = 3.6TSDD56 pKa = 2.77TTTPNASATMTPDD69 pKa = 3.13TSDD72 pKa = 2.9TTIPDD77 pKa = 3.7TSATMTPDD85 pKa = 3.11TSDD88 pKa = 2.9TTIPDD93 pKa = 3.56TSDD96 pKa = 2.49TMTPDD101 pKa = 3.6PSDD104 pKa = 3.23TTTPNASATMTPDD117 pKa = 3.13TSDD120 pKa = 2.9TTIPDD125 pKa = 3.56TSDD128 pKa = 2.36TMTPDD133 pKa = 3.17TSDD136 pKa = 2.9TTIPDD141 pKa = 3.56TSDD144 pKa = 2.52TMTPNASATMTPDD157 pKa = 3.09TSDD160 pKa = 2.61TMTPDD165 pKa = 3.12TSDD168 pKa = 2.98TTTPNASATMTPDD181 pKa = 3.13TSDD184 pKa = 2.9TTIPDD189 pKa = 3.6TSDD192 pKa = 2.75TTTPNVSATMTPDD205 pKa = 3.15TSDD208 pKa = 2.9TTIPDD213 pKa = 3.6TSDD216 pKa = 2.77TTTPNASATMTPDD229 pKa = 3.13TSDD232 pKa = 2.9TTIPDD237 pKa = 3.6TSDD240 pKa = 2.77TTTPNASATMTPDD253 pKa = 3.13TSDD256 pKa = 2.9TTIPDD261 pKa = 3.6TSDD264 pKa = 2.77TTTPNASATMTPDD277 pKa = 3.13TSDD280 pKa = 2.9TTIPDD285 pKa = 3.6TSDD288 pKa = 2.75TTTPNVSATMTPDD301 pKa = 3.15TSDD304 pKa = 2.9TTIPDD309 pKa = 3.6TSDD312 pKa = 2.77TTTPNASATMTPDD325 pKa = 3.13TSDD328 pKa = 2.9TTIPDD333 pKa = 3.6TSDD336 pKa = 2.77TTTPNASATMTPDD349 pKa = 3.13TSDD352 pKa = 2.9TTIPDD357 pKa = 3.6TSDD360 pKa = 2.75TTTPNVSATMTPDD373 pKa = 3.15TSDD376 pKa = 2.9TTIPDD381 pKa = 3.6TSDD384 pKa = 2.75TTTPNVSATMTPDD397 pKa = 3.15TSDD400 pKa = 2.9TTIPDD405 pKa = 3.56TSDD408 pKa = 2.52TMTPNASATMTPDD421 pKa = 3.13TSDD424 pKa = 2.9TTIPDD429 pKa = 3.6TSDD432 pKa = 2.77TTTPNASATMTPDD445 pKa = 3.15TSDD448 pKa = 3.08TTILDD453 pKa = 3.76TSATMTPDD461 pKa = 3.08TSDD464 pKa = 2.99TTTPEE469 pKa = 3.91PSATTTPDD477 pKa = 3.38PFDD480 pKa = 3.55TTTPDD485 pKa = 3.34PSATTTLDD493 pKa = 3.54PSDD496 pKa = 3.85TTTPDD501 pKa = 3.22PSATTTPDD509 pKa = 2.86SSTLTTPDD517 pKa = 3.73PSAMTTPDD525 pKa = 3.82PSTVTLDD532 pKa = 3.76PTNSTPTLDD541 pKa = 3.83PTTLTATPTSSSPASTPDD559 pKa = 3.0SSTSALTPTSSVSTSTLTSSVTPPTPDD586 pKa = 3.18SPSTPTSSIPTTTNITGSSALTTTSSTAPDD616 pKa = 3.31ISSTPTASSAATTSLVTTTTPSTTTSPTDD645 pKa = 3.06VTTTINSPMTTSGNTTSPTTTTMMTTGATNTTTLPTTTTTTTKK688 pKa = 9.18PTTTTTHH695 pKa = 5.52STTTTTTTTTTTTTTTTSTMSPTTTPTTTTPGVCEE730 pKa = 4.78NGGTWVNGQCVCPPGYY746 pKa = 9.2TGNTCSSIEE755 pKa = 3.98SSIEE759 pKa = 3.79TKK761 pKa = 10.92AEE763 pKa = 4.11TNVSVSVEE771 pKa = 3.68LRR773 pKa = 11.84VTNQEE778 pKa = 3.7FTEE781 pKa = 4.07DD782 pKa = 3.69LRR784 pKa = 11.84NEE786 pKa = 3.86SSKK789 pKa = 10.81GYY791 pKa = 10.33KK792 pKa = 10.13SFVLTFTLQMEE803 pKa = 4.68IYY805 pKa = 8.72YY806 pKa = 10.5RR807 pKa = 11.84NIEE810 pKa = 4.24GYY812 pKa = 9.96QGIKK816 pKa = 9.84ILSLSPGSIVVNHH829 pKa = 5.38TVIVAVPVTADD840 pKa = 3.34TVEE843 pKa = 4.44KK844 pKa = 10.35IGNVTKK850 pKa = 10.73NVEE853 pKa = 4.05KK854 pKa = 10.72EE855 pKa = 3.97IVQAAAQHH863 pKa = 5.63EE864 pKa = 4.67NCTEE868 pKa = 3.68NSTFCFNASEE878 pKa = 4.22VTIAQPSYY886 pKa = 9.96EE887 pKa = 4.28FNATEE892 pKa = 4.34YY893 pKa = 10.46CRR895 pKa = 11.84QVTPNGFKK903 pKa = 10.73DD904 pKa = 4.34FYY906 pKa = 11.33YY907 pKa = 10.45PNTTKK912 pKa = 10.9NGLTCVSNCSANTASAIDD930 pKa = 4.15CNEE933 pKa = 3.99GQCQLTLRR941 pKa = 11.84GPQCFCRR948 pKa = 11.84EE949 pKa = 4.08TNLYY953 pKa = 8.61WYY955 pKa = 10.49QGDD958 pKa = 3.73RR959 pKa = 11.84CSSRR963 pKa = 11.84VSKK966 pKa = 10.08MAVGMGLAVAVLCLVIFGLAAFIFRR991 pKa = 11.84ARR993 pKa = 11.84RR994 pKa = 11.84LKK996 pKa = 11.03LSDD999 pKa = 3.71RR1000 pKa = 11.84YY1001 pKa = 11.06SDD1003 pKa = 3.43LKK1005 pKa = 9.11EE1006 pKa = 4.46TKK1008 pKa = 8.36EE1009 pKa = 3.61QWYY1012 pKa = 10.27EE1013 pKa = 3.92EE1014 pKa = 4.1EE1015 pKa = 4.43DD1016 pKa = 4.27EE1017 pKa = 4.17NWKK1020 pKa = 10.91APGGFTIQNDD1030 pKa = 4.32GAHH1033 pKa = 7.05EE1034 pKa = 3.94DD1035 pKa = 3.6TMRR1038 pKa = 11.84VDD1040 pKa = 4.0LQSVDD1045 pKa = 3.36TSAKK1049 pKa = 9.78VPTSAATATSLSTNSTTMVPNTTTTMSPTTSINSSLPTTSNLTTSITSSPTSASSPTTISRR1110 pKa = 11.84PNTTTTRR1117 pKa = 11.84PTTTTTTTTTTTITTTTTTTPGVCEE1142 pKa = 4.78NGGTWVNGQCVCLPGYY1158 pKa = 9.26TGTTCSYY1165 pKa = 9.52VGSSIEE1171 pKa = 4.23TKK1173 pKa = 10.8AEE1175 pKa = 4.11TNVSVSVEE1183 pKa = 3.59LRR1185 pKa = 11.84VTNRR1189 pKa = 11.84EE1190 pKa = 3.67FTEE1193 pKa = 3.97DD1194 pKa = 3.27LRR1196 pKa = 11.84NEE1198 pKa = 3.71NSEE1201 pKa = 4.31GYY1203 pKa = 10.36KK1204 pKa = 10.54SFVHH1208 pKa = 6.32NFTLQMEE1215 pKa = 4.79IYY1217 pKa = 8.72YY1218 pKa = 10.5RR1219 pKa = 11.84NIEE1222 pKa = 4.24GYY1224 pKa = 9.96QGIKK1228 pKa = 9.85ILSLSNGSIVVNHH1241 pKa = 5.39TVIVAVPVTTDD1252 pKa = 3.16TVKK1255 pKa = 10.95KK1256 pKa = 9.77IDD1258 pKa = 3.75NVTKK1262 pKa = 10.15NVEE1265 pKa = 4.37EE1266 pKa = 4.16IVQAAAQQEE1275 pKa = 4.42NCTDD1279 pKa = 3.1NSTFCFNASEE1289 pKa = 4.22VTIAQPSYY1297 pKa = 10.22EE1298 pKa = 4.41FNATAYY1304 pKa = 8.7CHH1306 pKa = 5.75QVAPSDD1312 pKa = 3.96FKK1314 pKa = 11.46DD1315 pKa = 4.51FYY1317 pKa = 10.92YY1318 pKa = 10.47PNITKK1323 pKa = 10.5SGLTCVSNCSANTASAIDD1341 pKa = 4.15CNEE1344 pKa = 3.92GQCQLTHH1351 pKa = 6.91NGPQCLL1357 pKa = 3.73
MM1 pKa = 7.4TPNPSTTTTPDD12 pKa = 2.66ISTSTLVPSASTLTTPEE29 pKa = 3.57SSYY32 pKa = 9.68PTTPDD37 pKa = 2.87TSATTIPDD45 pKa = 3.56TSDD48 pKa = 2.73TTIPDD53 pKa = 3.6TSDD56 pKa = 2.77TTTPNASATMTPDD69 pKa = 3.13TSDD72 pKa = 2.9TTIPDD77 pKa = 3.7TSATMTPDD85 pKa = 3.11TSDD88 pKa = 2.9TTIPDD93 pKa = 3.56TSDD96 pKa = 2.49TMTPDD101 pKa = 3.6PSDD104 pKa = 3.23TTTPNASATMTPDD117 pKa = 3.13TSDD120 pKa = 2.9TTIPDD125 pKa = 3.56TSDD128 pKa = 2.36TMTPDD133 pKa = 3.17TSDD136 pKa = 2.9TTIPDD141 pKa = 3.56TSDD144 pKa = 2.52TMTPNASATMTPDD157 pKa = 3.09TSDD160 pKa = 2.61TMTPDD165 pKa = 3.12TSDD168 pKa = 2.98TTTPNASATMTPDD181 pKa = 3.13TSDD184 pKa = 2.9TTIPDD189 pKa = 3.6TSDD192 pKa = 2.75TTTPNVSATMTPDD205 pKa = 3.15TSDD208 pKa = 2.9TTIPDD213 pKa = 3.6TSDD216 pKa = 2.77TTTPNASATMTPDD229 pKa = 3.13TSDD232 pKa = 2.9TTIPDD237 pKa = 3.6TSDD240 pKa = 2.77TTTPNASATMTPDD253 pKa = 3.13TSDD256 pKa = 2.9TTIPDD261 pKa = 3.6TSDD264 pKa = 2.77TTTPNASATMTPDD277 pKa = 3.13TSDD280 pKa = 2.9TTIPDD285 pKa = 3.6TSDD288 pKa = 2.75TTTPNVSATMTPDD301 pKa = 3.15TSDD304 pKa = 2.9TTIPDD309 pKa = 3.6TSDD312 pKa = 2.77TTTPNASATMTPDD325 pKa = 3.13TSDD328 pKa = 2.9TTIPDD333 pKa = 3.6TSDD336 pKa = 2.77TTTPNASATMTPDD349 pKa = 3.13TSDD352 pKa = 2.9TTIPDD357 pKa = 3.6TSDD360 pKa = 2.75TTTPNVSATMTPDD373 pKa = 3.15TSDD376 pKa = 2.9TTIPDD381 pKa = 3.6TSDD384 pKa = 2.75TTTPNVSATMTPDD397 pKa = 3.15TSDD400 pKa = 2.9TTIPDD405 pKa = 3.56TSDD408 pKa = 2.52TMTPNASATMTPDD421 pKa = 3.13TSDD424 pKa = 2.9TTIPDD429 pKa = 3.6TSDD432 pKa = 2.77TTTPNASATMTPDD445 pKa = 3.15TSDD448 pKa = 3.08TTILDD453 pKa = 3.76TSATMTPDD461 pKa = 3.08TSDD464 pKa = 2.99TTTPEE469 pKa = 3.91PSATTTPDD477 pKa = 3.38PFDD480 pKa = 3.55TTTPDD485 pKa = 3.34PSATTTLDD493 pKa = 3.54PSDD496 pKa = 3.85TTTPDD501 pKa = 3.22PSATTTPDD509 pKa = 2.86SSTLTTPDD517 pKa = 3.73PSAMTTPDD525 pKa = 3.82PSTVTLDD532 pKa = 3.76PTNSTPTLDD541 pKa = 3.83PTTLTATPTSSSPASTPDD559 pKa = 3.0SSTSALTPTSSVSTSTLTSSVTPPTPDD586 pKa = 3.18SPSTPTSSIPTTTNITGSSALTTTSSTAPDD616 pKa = 3.31ISSTPTASSAATTSLVTTTTPSTTTSPTDD645 pKa = 3.06VTTTINSPMTTSGNTTSPTTTTMMTTGATNTTTLPTTTTTTTKK688 pKa = 9.18PTTTTTHH695 pKa = 5.52STTTTTTTTTTTTTTTTSTMSPTTTPTTTTPGVCEE730 pKa = 4.78NGGTWVNGQCVCPPGYY746 pKa = 9.2TGNTCSSIEE755 pKa = 3.98SSIEE759 pKa = 3.79TKK761 pKa = 10.92AEE763 pKa = 4.11TNVSVSVEE771 pKa = 3.68LRR773 pKa = 11.84VTNQEE778 pKa = 3.7FTEE781 pKa = 4.07DD782 pKa = 3.69LRR784 pKa = 11.84NEE786 pKa = 3.86SSKK789 pKa = 10.81GYY791 pKa = 10.33KK792 pKa = 10.13SFVLTFTLQMEE803 pKa = 4.68IYY805 pKa = 8.72YY806 pKa = 10.5RR807 pKa = 11.84NIEE810 pKa = 4.24GYY812 pKa = 9.96QGIKK816 pKa = 9.84ILSLSPGSIVVNHH829 pKa = 5.38TVIVAVPVTADD840 pKa = 3.34TVEE843 pKa = 4.44KK844 pKa = 10.35IGNVTKK850 pKa = 10.73NVEE853 pKa = 4.05KK854 pKa = 10.72EE855 pKa = 3.97IVQAAAQHH863 pKa = 5.63EE864 pKa = 4.67NCTEE868 pKa = 3.68NSTFCFNASEE878 pKa = 4.22VTIAQPSYY886 pKa = 9.96EE887 pKa = 4.28FNATEE892 pKa = 4.34YY893 pKa = 10.46CRR895 pKa = 11.84QVTPNGFKK903 pKa = 10.73DD904 pKa = 4.34FYY906 pKa = 11.33YY907 pKa = 10.45PNTTKK912 pKa = 10.9NGLTCVSNCSANTASAIDD930 pKa = 4.15CNEE933 pKa = 3.99GQCQLTLRR941 pKa = 11.84GPQCFCRR948 pKa = 11.84EE949 pKa = 4.08TNLYY953 pKa = 8.61WYY955 pKa = 10.49QGDD958 pKa = 3.73RR959 pKa = 11.84CSSRR963 pKa = 11.84VSKK966 pKa = 10.08MAVGMGLAVAVLCLVIFGLAAFIFRR991 pKa = 11.84ARR993 pKa = 11.84RR994 pKa = 11.84LKK996 pKa = 11.03LSDD999 pKa = 3.71RR1000 pKa = 11.84YY1001 pKa = 11.06SDD1003 pKa = 3.43LKK1005 pKa = 9.11EE1006 pKa = 4.46TKK1008 pKa = 8.36EE1009 pKa = 3.61QWYY1012 pKa = 10.27EE1013 pKa = 3.92EE1014 pKa = 4.1EE1015 pKa = 4.43DD1016 pKa = 4.27EE1017 pKa = 4.17NWKK1020 pKa = 10.91APGGFTIQNDD1030 pKa = 4.32GAHH1033 pKa = 7.05EE1034 pKa = 3.94DD1035 pKa = 3.6TMRR1038 pKa = 11.84VDD1040 pKa = 4.0LQSVDD1045 pKa = 3.36TSAKK1049 pKa = 9.78VPTSAATATSLSTNSTTMVPNTTTTMSPTTSINSSLPTTSNLTTSITSSPTSASSPTTISRR1110 pKa = 11.84PNTTTTRR1117 pKa = 11.84PTTTTTTTTTTTITTTTTTTPGVCEE1142 pKa = 4.78NGGTWVNGQCVCLPGYY1158 pKa = 9.26TGTTCSYY1165 pKa = 9.52VGSSIEE1171 pKa = 4.23TKK1173 pKa = 10.8AEE1175 pKa = 4.11TNVSVSVEE1183 pKa = 3.59LRR1185 pKa = 11.84VTNRR1189 pKa = 11.84EE1190 pKa = 3.67FTEE1193 pKa = 3.97DD1194 pKa = 3.27LRR1196 pKa = 11.84NEE1198 pKa = 3.71NSEE1201 pKa = 4.31GYY1203 pKa = 10.36KK1204 pKa = 10.54SFVHH1208 pKa = 6.32NFTLQMEE1215 pKa = 4.79IYY1217 pKa = 8.72YY1218 pKa = 10.5RR1219 pKa = 11.84NIEE1222 pKa = 4.24GYY1224 pKa = 9.96QGIKK1228 pKa = 9.85ILSLSNGSIVVNHH1241 pKa = 5.39TVIVAVPVTTDD1252 pKa = 3.16TVKK1255 pKa = 10.95KK1256 pKa = 9.77IDD1258 pKa = 3.75NVTKK1262 pKa = 10.15NVEE1265 pKa = 4.37EE1266 pKa = 4.16IVQAAAQQEE1275 pKa = 4.42NCTDD1279 pKa = 3.1NSTFCFNASEE1289 pKa = 4.22VTIAQPSYY1297 pKa = 10.22EE1298 pKa = 4.41FNATAYY1304 pKa = 8.7CHH1306 pKa = 5.75QVAPSDD1312 pKa = 3.96FKK1314 pKa = 11.46DD1315 pKa = 4.51FYY1317 pKa = 10.92YY1318 pKa = 10.47PNITKK1323 pKa = 10.5SGLTCVSNCSANTASAIDD1341 pKa = 4.15CNEE1344 pKa = 3.92GQCQLTHH1351 pKa = 6.91NGPQCLL1357 pKa = 3.73
Molecular weight: 141.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J3DL10|A0A6J3DL10_AYTFU small integral membrane protein 4 OS=Aythya fuligula OX=219594 GN=SMIM4 PE=4 SV=1
MM1 pKa = 7.57SSHH4 pKa = 6.01KK5 pKa = 9.07TFKK8 pKa = 10.28IKK10 pKa = 10.6RR11 pKa = 11.84FLAKK15 pKa = 9.68KK16 pKa = 9.58QKK18 pKa = 8.69QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84MKK30 pKa = 9.89TGNKK34 pKa = 8.61IRR36 pKa = 11.84YY37 pKa = 7.09NSKK40 pKa = 8.3RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 3.95WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
MM1 pKa = 7.57SSHH4 pKa = 6.01KK5 pKa = 9.07TFKK8 pKa = 10.28IKK10 pKa = 10.6RR11 pKa = 11.84FLAKK15 pKa = 9.68KK16 pKa = 9.58QKK18 pKa = 8.69QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84MKK30 pKa = 9.89TGNKK34 pKa = 8.61IRR36 pKa = 11.84YY37 pKa = 7.09NSKK40 pKa = 8.3RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 3.95WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
Molecular weight: 6.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17668086 |
31 |
33862 |
735.1 |
82.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.888 ± 0.016 | 2.05 ± 0.012 |
5.066 ± 0.009 | 7.48 ± 0.021 |
3.459 ± 0.01 | 6.142 ± 0.023 |
2.477 ± 0.006 | 4.557 ± 0.012 |
6.214 ± 0.02 | 9.401 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.007 | 3.968 ± 0.013 |
5.834 ± 0.022 | 4.897 ± 0.016 |
5.467 ± 0.014 | 8.603 ± 0.022 |
5.438 ± 0.014 | 6.031 ± 0.014 |
1.08 ± 0.005 | 2.731 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
