
Hanstruepera neustonica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Hanstruepera
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
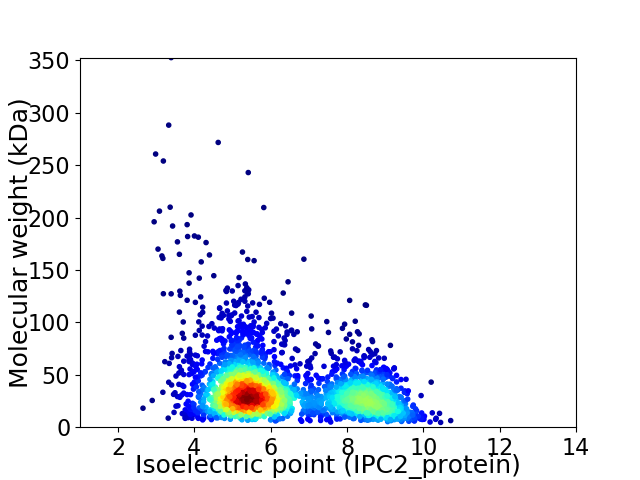
Virtual 2D-PAGE plot for 2739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K1DWZ9|A0A2K1DWZ9_9FLAO Uncharacterized protein OS=Hanstruepera neustonica OX=1445657 GN=C1T31_10450 PE=4 SV=1
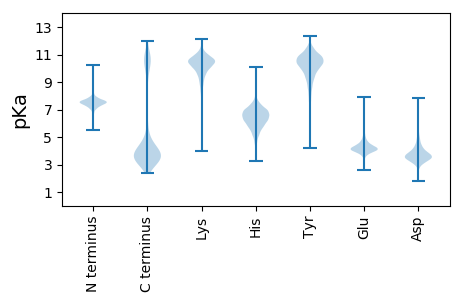
MM1 pKa = 7.34RR2 pKa = 11.84QIGLLLFVFFTGLLQAQISFEE23 pKa = 4.19DD24 pKa = 3.6QASDD28 pKa = 4.13LGVQLSCGTTYY39 pKa = 11.02LGNGVSFYY47 pKa = 11.4DD48 pKa = 4.25FDD50 pKa = 4.65NDD52 pKa = 3.36GWDD55 pKa = 3.89DD56 pKa = 3.67LTLNTEE62 pKa = 4.68FGQEE66 pKa = 3.19IRR68 pKa = 11.84FFKK71 pKa = 9.2NTNGTFAEE79 pKa = 4.31IFPTIPYY86 pKa = 9.77FDD88 pKa = 4.24YY89 pKa = 9.74QTKK92 pKa = 8.93QVNWVDD98 pKa = 4.07FDD100 pKa = 4.14NDD102 pKa = 3.2GDD104 pKa = 3.89KK105 pKa = 11.47DD106 pKa = 4.15LFVTSDD112 pKa = 3.54YY113 pKa = 11.27EE114 pKa = 4.52GNRR117 pKa = 11.84LFEE120 pKa = 4.54NTGSMTFVDD129 pKa = 4.16ISVSSGIDD137 pKa = 3.14TTNEE141 pKa = 3.44FSYY144 pKa = 9.09GTSWGDD150 pKa = 3.53YY151 pKa = 10.93NNDD154 pKa = 3.11GYY156 pKa = 11.84LDD158 pKa = 3.45IFVSNRR164 pKa = 11.84TALHH168 pKa = 5.77NNILYY173 pKa = 10.46RR174 pKa = 11.84NNGDD178 pKa = 3.49GTFTDD183 pKa = 3.51VTLAAGLSGSGSGSFCSAFIDD204 pKa = 3.69INNDD208 pKa = 2.75GFQDD212 pKa = 3.66IYY214 pKa = 10.8ISNDD218 pKa = 2.87RR219 pKa = 11.84PVNPNILYY227 pKa = 10.46KK228 pKa = 11.13NNGDD232 pKa = 3.67GTFTDD237 pKa = 4.23ISASSGSDD245 pKa = 3.2VTIDD249 pKa = 3.24AMTVTVADD257 pKa = 4.54FNSDD261 pKa = 2.89GWFDD265 pKa = 3.4IYY267 pKa = 10.01ITNGPIGNVLLKK279 pKa = 11.16NNGNEE284 pKa = 4.15TFTDD288 pKa = 3.34IAVSSGTVFNSISWGASFFDD308 pKa = 4.87AEE310 pKa = 4.15NDD312 pKa = 3.37MDD314 pKa = 4.02QDD316 pKa = 4.46LYY318 pKa = 11.83VCGEE322 pKa = 4.17FDD324 pKa = 4.24ANQTNFLLAAFYY336 pKa = 10.6TNNDD340 pKa = 3.52DD341 pKa = 3.85EE342 pKa = 4.89TFNLSNEE349 pKa = 4.38CFPDD353 pKa = 3.78DD354 pKa = 3.61NRR356 pKa = 11.84AAYY359 pKa = 9.93SSAVGDD365 pKa = 3.96VDD367 pKa = 3.96NDD369 pKa = 4.14GYY371 pKa = 10.43PDD373 pKa = 5.41LIVTNSFDD381 pKa = 3.85QDD383 pKa = 2.86IFFWKK388 pKa = 8.56NTTPQTNNWVKK399 pKa = 11.02LEE401 pKa = 4.07LEE403 pKa = 4.41GTVSNRR409 pKa = 11.84DD410 pKa = 3.46GIGSVIEE417 pKa = 3.86VSVNGEE423 pKa = 3.5KK424 pKa = 10.39QYY426 pKa = 11.44AYY428 pKa = 6.82THH430 pKa = 6.43CGEE433 pKa = 5.26GYY435 pKa = 10.58LSQNSATKK443 pKa = 10.16IIGTGTHH450 pKa = 4.88TTIDD454 pKa = 4.01YY455 pKa = 10.97IKK457 pKa = 9.03VTWLSGIVDD466 pKa = 3.83ILYY469 pKa = 9.84DD470 pKa = 3.51VPVNQKK476 pKa = 9.43IDD478 pKa = 3.76LVEE481 pKa = 4.14GTFPEE486 pKa = 4.51LSVAEE491 pKa = 4.16YY492 pKa = 10.91DD493 pKa = 3.85LQNTEE498 pKa = 5.14LYY500 pKa = 9.31PNPVNDD506 pKa = 3.93NLVVAVNEE514 pKa = 4.1DD515 pKa = 3.37ARR517 pKa = 11.84VEE519 pKa = 4.02IYY521 pKa = 10.24TILGKK526 pKa = 10.83RR527 pKa = 11.84MMTTNVSGQGNNQLDD542 pKa = 4.21VSTLPSGMYY551 pKa = 9.52LVKK554 pKa = 10.41ISADD558 pKa = 3.11NGQTLTRR565 pKa = 11.84KK566 pKa = 9.83LIKK569 pKa = 10.21KK570 pKa = 9.67
MM1 pKa = 7.34RR2 pKa = 11.84QIGLLLFVFFTGLLQAQISFEE23 pKa = 4.19DD24 pKa = 3.6QASDD28 pKa = 4.13LGVQLSCGTTYY39 pKa = 11.02LGNGVSFYY47 pKa = 11.4DD48 pKa = 4.25FDD50 pKa = 4.65NDD52 pKa = 3.36GWDD55 pKa = 3.89DD56 pKa = 3.67LTLNTEE62 pKa = 4.68FGQEE66 pKa = 3.19IRR68 pKa = 11.84FFKK71 pKa = 9.2NTNGTFAEE79 pKa = 4.31IFPTIPYY86 pKa = 9.77FDD88 pKa = 4.24YY89 pKa = 9.74QTKK92 pKa = 8.93QVNWVDD98 pKa = 4.07FDD100 pKa = 4.14NDD102 pKa = 3.2GDD104 pKa = 3.89KK105 pKa = 11.47DD106 pKa = 4.15LFVTSDD112 pKa = 3.54YY113 pKa = 11.27EE114 pKa = 4.52GNRR117 pKa = 11.84LFEE120 pKa = 4.54NTGSMTFVDD129 pKa = 4.16ISVSSGIDD137 pKa = 3.14TTNEE141 pKa = 3.44FSYY144 pKa = 9.09GTSWGDD150 pKa = 3.53YY151 pKa = 10.93NNDD154 pKa = 3.11GYY156 pKa = 11.84LDD158 pKa = 3.45IFVSNRR164 pKa = 11.84TALHH168 pKa = 5.77NNILYY173 pKa = 10.46RR174 pKa = 11.84NNGDD178 pKa = 3.49GTFTDD183 pKa = 3.51VTLAAGLSGSGSGSFCSAFIDD204 pKa = 3.69INNDD208 pKa = 2.75GFQDD212 pKa = 3.66IYY214 pKa = 10.8ISNDD218 pKa = 2.87RR219 pKa = 11.84PVNPNILYY227 pKa = 10.46KK228 pKa = 11.13NNGDD232 pKa = 3.67GTFTDD237 pKa = 4.23ISASSGSDD245 pKa = 3.2VTIDD249 pKa = 3.24AMTVTVADD257 pKa = 4.54FNSDD261 pKa = 2.89GWFDD265 pKa = 3.4IYY267 pKa = 10.01ITNGPIGNVLLKK279 pKa = 11.16NNGNEE284 pKa = 4.15TFTDD288 pKa = 3.34IAVSSGTVFNSISWGASFFDD308 pKa = 4.87AEE310 pKa = 4.15NDD312 pKa = 3.37MDD314 pKa = 4.02QDD316 pKa = 4.46LYY318 pKa = 11.83VCGEE322 pKa = 4.17FDD324 pKa = 4.24ANQTNFLLAAFYY336 pKa = 10.6TNNDD340 pKa = 3.52DD341 pKa = 3.85EE342 pKa = 4.89TFNLSNEE349 pKa = 4.38CFPDD353 pKa = 3.78DD354 pKa = 3.61NRR356 pKa = 11.84AAYY359 pKa = 9.93SSAVGDD365 pKa = 3.96VDD367 pKa = 3.96NDD369 pKa = 4.14GYY371 pKa = 10.43PDD373 pKa = 5.41LIVTNSFDD381 pKa = 3.85QDD383 pKa = 2.86IFFWKK388 pKa = 8.56NTTPQTNNWVKK399 pKa = 11.02LEE401 pKa = 4.07LEE403 pKa = 4.41GTVSNRR409 pKa = 11.84DD410 pKa = 3.46GIGSVIEE417 pKa = 3.86VSVNGEE423 pKa = 3.5KK424 pKa = 10.39QYY426 pKa = 11.44AYY428 pKa = 6.82THH430 pKa = 6.43CGEE433 pKa = 5.26GYY435 pKa = 10.58LSQNSATKK443 pKa = 10.16IIGTGTHH450 pKa = 4.88TTIDD454 pKa = 4.01YY455 pKa = 10.97IKK457 pKa = 9.03VTWLSGIVDD466 pKa = 3.83ILYY469 pKa = 9.84DD470 pKa = 3.51VPVNQKK476 pKa = 9.43IDD478 pKa = 3.76LVEE481 pKa = 4.14GTFPEE486 pKa = 4.51LSVAEE491 pKa = 4.16YY492 pKa = 10.91DD493 pKa = 3.85LQNTEE498 pKa = 5.14LYY500 pKa = 9.31PNPVNDD506 pKa = 3.93NLVVAVNEE514 pKa = 4.1DD515 pKa = 3.37ARR517 pKa = 11.84VEE519 pKa = 4.02IYY521 pKa = 10.24TILGKK526 pKa = 10.83RR527 pKa = 11.84MMTTNVSGQGNNQLDD542 pKa = 4.21VSTLPSGMYY551 pKa = 9.52LVKK554 pKa = 10.41ISADD558 pKa = 3.11NGQTLTRR565 pKa = 11.84KK566 pKa = 9.83LIKK569 pKa = 10.21KK570 pKa = 9.67
Molecular weight: 62.98 kDa
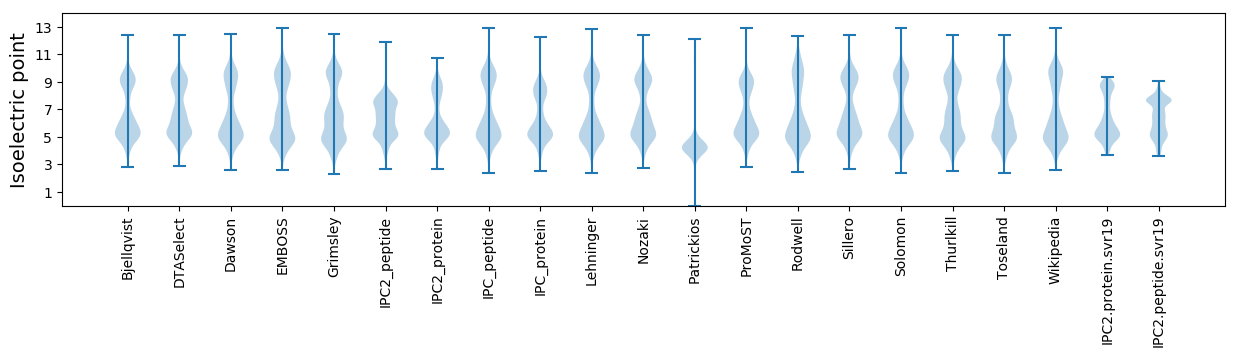
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K1DWP2|A0A2K1DWP2_9FLAO Glycosyltransferase OS=Hanstruepera neustonica OX=1445657 GN=C1T31_11690 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.16RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.52LSVSSEE48 pKa = 3.87PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 10.15RR11 pKa = 11.84KK12 pKa = 9.6RR13 pKa = 11.84KK14 pKa = 8.29NKK16 pKa = 9.34HH17 pKa = 4.03GFRR20 pKa = 11.84EE21 pKa = 4.16RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.52LSVSSEE48 pKa = 3.87PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
942191 |
38 |
3366 |
344.0 |
38.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.205 ± 0.046 | 0.815 ± 0.024 |
5.984 ± 0.051 | 6.411 ± 0.052 |
5.23 ± 0.041 | 6.369 ± 0.06 |
1.826 ± 0.021 | 7.957 ± 0.043 |
7.21 ± 0.078 | 9.13 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.026 | 6.469 ± 0.049 |
3.427 ± 0.034 | 3.483 ± 0.029 |
3.254 ± 0.031 | 6.537 ± 0.039 |
5.959 ± 0.071 | 6.362 ± 0.037 |
1.023 ± 0.017 | 4.116 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
