
Psychromonas sp. RZ5
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Psychromonadaceae; Psychromonas; unclassified Psychromonas
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
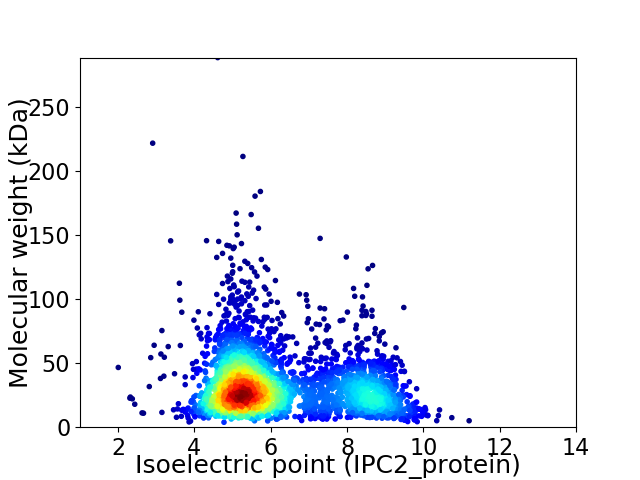
Virtual 2D-PAGE plot for 3176 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y7ZIL1|A0A4Y7ZIL1_9GAMM 1 4-dihydroxy-2-naphthoate octaprenyltransferase OS=Psychromonas sp. RZ5 OX=2555642 GN=menA PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.3KK3 pKa = 10.46ALLAVLIPSLLGATSAFAGGVDD25 pKa = 5.4LIKK28 pKa = 10.95NDD30 pKa = 4.68DD31 pKa = 3.42MTLNFNGDD39 pKa = 3.49IDD41 pKa = 4.43LKK43 pKa = 11.25SYY45 pKa = 8.97ITFTDD50 pKa = 2.87TDD52 pKa = 3.54GDD54 pKa = 4.04YY55 pKa = 11.04KK56 pKa = 11.35SNSGEE61 pKa = 4.13TEE63 pKa = 4.15VIFDD67 pKa = 5.38DD68 pKa = 5.01IDD70 pKa = 4.85FKK72 pKa = 10.45FTWHH76 pKa = 7.05ASDD79 pKa = 3.67NLDD82 pKa = 4.18YY83 pKa = 10.92IAGMDD88 pKa = 4.12LTADD92 pKa = 3.62PDD94 pKa = 4.21GYY96 pKa = 9.43TEE98 pKa = 4.02IGAAVGVFYY107 pKa = 10.77AWAGFNHH114 pKa = 5.84SQYY117 pKa = 11.55GQLTWGNQITSFDD130 pKa = 3.54NFGIDD135 pKa = 3.33TAEE138 pKa = 3.68IWGGAASGKK147 pKa = 10.17LDD149 pKa = 3.47GRR151 pKa = 11.84STEE154 pKa = 3.82HH155 pKa = 6.91SNALVYY161 pKa = 10.42EE162 pKa = 4.05IEE164 pKa = 4.57IDD166 pKa = 3.91DD167 pKa = 3.99LTISATYY174 pKa = 10.01GINNEE179 pKa = 4.61DD180 pKa = 4.91DD181 pKa = 4.35SDD183 pKa = 3.95DD184 pKa = 3.88FKK186 pKa = 11.27NAKK189 pKa = 8.96VMQVTAFYY197 pKa = 10.81KK198 pKa = 10.76PGDD201 pKa = 3.69LQINAGVGHH210 pKa = 4.91TTGYY214 pKa = 9.83NQDD217 pKa = 3.56SADD220 pKa = 3.61FAQVSTAVYY229 pKa = 8.68GQAEE233 pKa = 4.49VEE235 pKa = 4.24YY236 pKa = 10.52QMGDD240 pKa = 2.72ISVAGLVAYY249 pKa = 10.29QDD251 pKa = 3.32AAYY254 pKa = 9.68EE255 pKa = 4.26DD256 pKa = 4.3NSANTGSDD264 pKa = 3.45FTSTGVEE271 pKa = 3.63FDD273 pKa = 3.37VTYY276 pKa = 10.94KK277 pKa = 10.85LSSTVKK283 pKa = 10.02LSVGGDD289 pKa = 3.98YY290 pKa = 9.6ITQDD294 pKa = 2.86IDD296 pKa = 3.61NVSDD300 pKa = 3.21NDD302 pKa = 3.65AYY304 pKa = 10.88MDD306 pKa = 4.14LFVASKK312 pKa = 8.72HH313 pKa = 4.89TLTKK317 pKa = 10.45NVYY320 pKa = 10.19LLTEE324 pKa = 4.41LAMQQGDD331 pKa = 4.37LIEE334 pKa = 4.1YY335 pKa = 10.05RR336 pKa = 11.84ADD338 pKa = 3.45KK339 pKa = 10.97DD340 pKa = 4.22SVSTTSLDD348 pKa = 3.57DD349 pKa = 3.99AQVKK353 pKa = 10.48VGFLLNMQFF362 pKa = 4.04
MM1 pKa = 7.52KK2 pKa = 10.3KK3 pKa = 10.46ALLAVLIPSLLGATSAFAGGVDD25 pKa = 5.4LIKK28 pKa = 10.95NDD30 pKa = 4.68DD31 pKa = 3.42MTLNFNGDD39 pKa = 3.49IDD41 pKa = 4.43LKK43 pKa = 11.25SYY45 pKa = 8.97ITFTDD50 pKa = 2.87TDD52 pKa = 3.54GDD54 pKa = 4.04YY55 pKa = 11.04KK56 pKa = 11.35SNSGEE61 pKa = 4.13TEE63 pKa = 4.15VIFDD67 pKa = 5.38DD68 pKa = 5.01IDD70 pKa = 4.85FKK72 pKa = 10.45FTWHH76 pKa = 7.05ASDD79 pKa = 3.67NLDD82 pKa = 4.18YY83 pKa = 10.92IAGMDD88 pKa = 4.12LTADD92 pKa = 3.62PDD94 pKa = 4.21GYY96 pKa = 9.43TEE98 pKa = 4.02IGAAVGVFYY107 pKa = 10.77AWAGFNHH114 pKa = 5.84SQYY117 pKa = 11.55GQLTWGNQITSFDD130 pKa = 3.54NFGIDD135 pKa = 3.33TAEE138 pKa = 3.68IWGGAASGKK147 pKa = 10.17LDD149 pKa = 3.47GRR151 pKa = 11.84STEE154 pKa = 3.82HH155 pKa = 6.91SNALVYY161 pKa = 10.42EE162 pKa = 4.05IEE164 pKa = 4.57IDD166 pKa = 3.91DD167 pKa = 3.99LTISATYY174 pKa = 10.01GINNEE179 pKa = 4.61DD180 pKa = 4.91DD181 pKa = 4.35SDD183 pKa = 3.95DD184 pKa = 3.88FKK186 pKa = 11.27NAKK189 pKa = 8.96VMQVTAFYY197 pKa = 10.81KK198 pKa = 10.76PGDD201 pKa = 3.69LQINAGVGHH210 pKa = 4.91TTGYY214 pKa = 9.83NQDD217 pKa = 3.56SADD220 pKa = 3.61FAQVSTAVYY229 pKa = 8.68GQAEE233 pKa = 4.49VEE235 pKa = 4.24YY236 pKa = 10.52QMGDD240 pKa = 2.72ISVAGLVAYY249 pKa = 10.29QDD251 pKa = 3.32AAYY254 pKa = 9.68EE255 pKa = 4.26DD256 pKa = 4.3NSANTGSDD264 pKa = 3.45FTSTGVEE271 pKa = 3.63FDD273 pKa = 3.37VTYY276 pKa = 10.94KK277 pKa = 10.85LSSTVKK283 pKa = 10.02LSVGGDD289 pKa = 3.98YY290 pKa = 9.6ITQDD294 pKa = 2.86IDD296 pKa = 3.61NVSDD300 pKa = 3.21NDD302 pKa = 3.65AYY304 pKa = 10.88MDD306 pKa = 4.14LFVASKK312 pKa = 8.72HH313 pKa = 4.89TLTKK317 pKa = 10.45NVYY320 pKa = 10.19LLTEE324 pKa = 4.41LAMQQGDD331 pKa = 4.37LIEE334 pKa = 4.1YY335 pKa = 10.05RR336 pKa = 11.84ADD338 pKa = 3.45KK339 pKa = 10.97DD340 pKa = 4.22SVSTTSLDD348 pKa = 3.57DD349 pKa = 3.99AQVKK353 pKa = 10.48VGFLLNMQFF362 pKa = 4.04
Molecular weight: 39.3 kDa
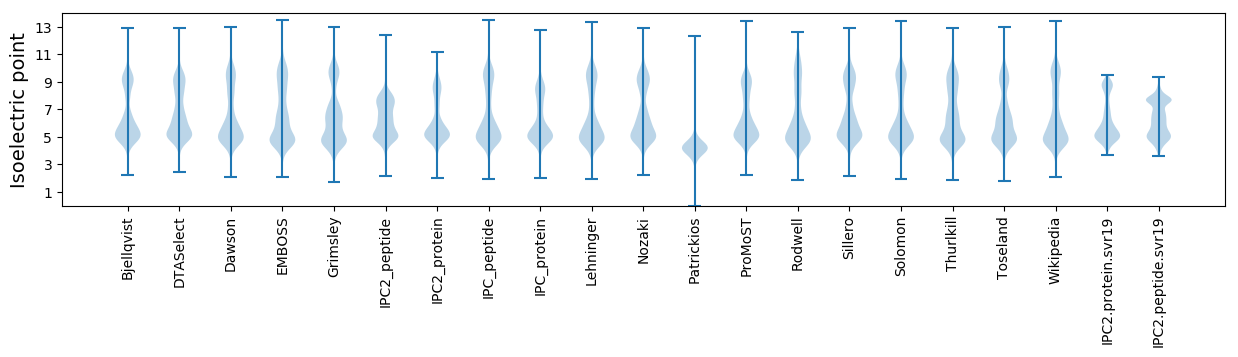
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y7ZMG5|A0A4Y7ZMG5_9GAMM Single-stranded-DNA-specific exonuclease RecJ OS=Psychromonas sp. RZ5 OX=2555642 GN=recJ PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNTKK11 pKa = 10.0RR12 pKa = 11.84KK13 pKa = 8.73RR14 pKa = 11.84SHH16 pKa = 6.16GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.18NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNTKK11 pKa = 10.0RR12 pKa = 11.84KK13 pKa = 8.73RR14 pKa = 11.84SHH16 pKa = 6.16GFRR19 pKa = 11.84TRR21 pKa = 11.84MATKK25 pKa = 10.18NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.5GRR39 pKa = 11.84ASLSAA44 pKa = 3.83
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992658 |
32 |
2661 |
312.5 |
34.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.078 ± 0.047 | 1.033 ± 0.015 |
5.45 ± 0.037 | 6.417 ± 0.049 |
4.422 ± 0.033 | 6.201 ± 0.046 |
2.098 ± 0.022 | 7.286 ± 0.038 |
6.218 ± 0.042 | 10.704 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.02 | 4.845 ± 0.036 |
3.497 ± 0.026 | 4.508 ± 0.041 |
3.778 ± 0.034 | 6.818 ± 0.044 |
5.436 ± 0.045 | 6.591 ± 0.036 |
1.087 ± 0.015 | 3.113 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
