
Ictalurid herpesvirus 1 (strain Auburn) (IcHV-1) (Channel catfish herpesvirus)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Alloherpesviridae; Ictalurivirus; Ictalurid herpesvirus 1
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 76 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|Q00115|VG03_ICHVA Uncharacterized protein ORF3 OS=Ictalurid herpesvirus 1 (strain Auburn) OX=766178 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.55SATQGPLTEE10 pKa = 4.73AANGCDD16 pKa = 3.4DD17 pKa = 3.86TGLGKK22 pKa = 8.7TKK24 pKa = 10.33NHH26 pKa = 6.59WDD28 pKa = 3.41GAVPRR33 pKa = 11.84TTGEE37 pKa = 3.89PRR39 pKa = 11.84SLLGEE44 pKa = 3.98NRR46 pKa = 11.84ASSVLTTGLGPKK58 pKa = 9.48PGPVWVDD65 pKa = 2.67PRR67 pKa = 11.84IRR69 pKa = 11.84FHH71 pKa = 6.76GVSEE75 pKa = 4.36SLPLTDD81 pKa = 4.8TPWGPPPSPMDD92 pKa = 3.98ALGHH96 pKa = 5.8GVMTPSGLKK105 pKa = 8.78WCPPPPPKK113 pKa = 10.17TEE115 pKa = 3.7SRR117 pKa = 11.84SFLEE121 pKa = 4.42LLAMHH126 pKa = 6.51QKK128 pKa = 10.33AQADD132 pKa = 3.83RR133 pKa = 11.84DD134 pKa = 3.96DD135 pKa = 4.09TVTGEE140 pKa = 4.38TEE142 pKa = 3.79TTARR146 pKa = 11.84EE147 pKa = 3.84EE148 pKa = 4.17DD149 pKa = 3.51DD150 pKa = 4.1VIFVEE155 pKa = 4.56EE156 pKa = 4.4TNVNNPDD163 pKa = 3.54VIEE166 pKa = 4.46VIEE169 pKa = 4.54LKK171 pKa = 10.83DD172 pKa = 3.53VTEE175 pKa = 4.56TGHH178 pKa = 5.5GTLKK182 pKa = 10.03RR183 pKa = 11.84RR184 pKa = 11.84YY185 pKa = 7.82PMRR188 pKa = 11.84VRR190 pKa = 11.84RR191 pKa = 11.84APKK194 pKa = 9.99RR195 pKa = 11.84LVLDD199 pKa = 3.73EE200 pKa = 4.48QVVDD204 pKa = 5.49DD205 pKa = 5.08YY206 pKa = 11.63PADD209 pKa = 3.99SDD211 pKa = 4.93DD212 pKa = 4.25DD213 pKa = 3.95TDD215 pKa = 4.99AEE217 pKa = 4.56SDD219 pKa = 3.57DD220 pKa = 3.93AMSVLSDD227 pKa = 3.39TCRR230 pKa = 11.84TDD232 pKa = 4.67DD233 pKa = 4.27DD234 pKa = 4.85ASSVSSCGSFITDD247 pKa = 3.61GSGSEE252 pKa = 4.17EE253 pKa = 4.57SEE255 pKa = 4.44DD256 pKa = 3.84SASDD260 pKa = 3.47EE261 pKa = 4.41TDD263 pKa = 4.19DD264 pKa = 5.59SDD266 pKa = 5.17FDD268 pKa = 4.12TDD270 pKa = 3.53EE271 pKa = 4.36LTSEE275 pKa = 4.28SEE277 pKa = 4.23EE278 pKa = 4.11EE279 pKa = 4.06EE280 pKa = 4.36SEE282 pKa = 4.39SEE284 pKa = 4.25SEE286 pKa = 4.37SEE288 pKa = 4.35SEE290 pKa = 4.55SEE292 pKa = 4.63SEE294 pKa = 4.76SEE296 pKa = 4.06
MM1 pKa = 7.55SATQGPLTEE10 pKa = 4.73AANGCDD16 pKa = 3.4DD17 pKa = 3.86TGLGKK22 pKa = 8.7TKK24 pKa = 10.33NHH26 pKa = 6.59WDD28 pKa = 3.41GAVPRR33 pKa = 11.84TTGEE37 pKa = 3.89PRR39 pKa = 11.84SLLGEE44 pKa = 3.98NRR46 pKa = 11.84ASSVLTTGLGPKK58 pKa = 9.48PGPVWVDD65 pKa = 2.67PRR67 pKa = 11.84IRR69 pKa = 11.84FHH71 pKa = 6.76GVSEE75 pKa = 4.36SLPLTDD81 pKa = 4.8TPWGPPPSPMDD92 pKa = 3.98ALGHH96 pKa = 5.8GVMTPSGLKK105 pKa = 8.78WCPPPPPKK113 pKa = 10.17TEE115 pKa = 3.7SRR117 pKa = 11.84SFLEE121 pKa = 4.42LLAMHH126 pKa = 6.51QKK128 pKa = 10.33AQADD132 pKa = 3.83RR133 pKa = 11.84DD134 pKa = 3.96DD135 pKa = 4.09TVTGEE140 pKa = 4.38TEE142 pKa = 3.79TTARR146 pKa = 11.84EE147 pKa = 3.84EE148 pKa = 4.17DD149 pKa = 3.51DD150 pKa = 4.1VIFVEE155 pKa = 4.56EE156 pKa = 4.4TNVNNPDD163 pKa = 3.54VIEE166 pKa = 4.46VIEE169 pKa = 4.54LKK171 pKa = 10.83DD172 pKa = 3.53VTEE175 pKa = 4.56TGHH178 pKa = 5.5GTLKK182 pKa = 10.03RR183 pKa = 11.84RR184 pKa = 11.84YY185 pKa = 7.82PMRR188 pKa = 11.84VRR190 pKa = 11.84RR191 pKa = 11.84APKK194 pKa = 9.99RR195 pKa = 11.84LVLDD199 pKa = 3.73EE200 pKa = 4.48QVVDD204 pKa = 5.49DD205 pKa = 5.08YY206 pKa = 11.63PADD209 pKa = 3.99SDD211 pKa = 4.93DD212 pKa = 4.25DD213 pKa = 3.95TDD215 pKa = 4.99AEE217 pKa = 4.56SDD219 pKa = 3.57DD220 pKa = 3.93AMSVLSDD227 pKa = 3.39TCRR230 pKa = 11.84TDD232 pKa = 4.67DD233 pKa = 4.27DD234 pKa = 4.85ASSVSSCGSFITDD247 pKa = 3.61GSGSEE252 pKa = 4.17EE253 pKa = 4.57SEE255 pKa = 4.44DD256 pKa = 3.84SASDD260 pKa = 3.47EE261 pKa = 4.41TDD263 pKa = 4.19DD264 pKa = 5.59SDD266 pKa = 5.17FDD268 pKa = 4.12TDD270 pKa = 3.53EE271 pKa = 4.36LTSEE275 pKa = 4.28SEE277 pKa = 4.23EE278 pKa = 4.11EE279 pKa = 4.06EE280 pKa = 4.36SEE282 pKa = 4.39SEE284 pKa = 4.25SEE286 pKa = 4.37SEE288 pKa = 4.35SEE290 pKa = 4.55SEE292 pKa = 4.63SEE294 pKa = 4.76SEE296 pKa = 4.06
Molecular weight: 32.0 kDa
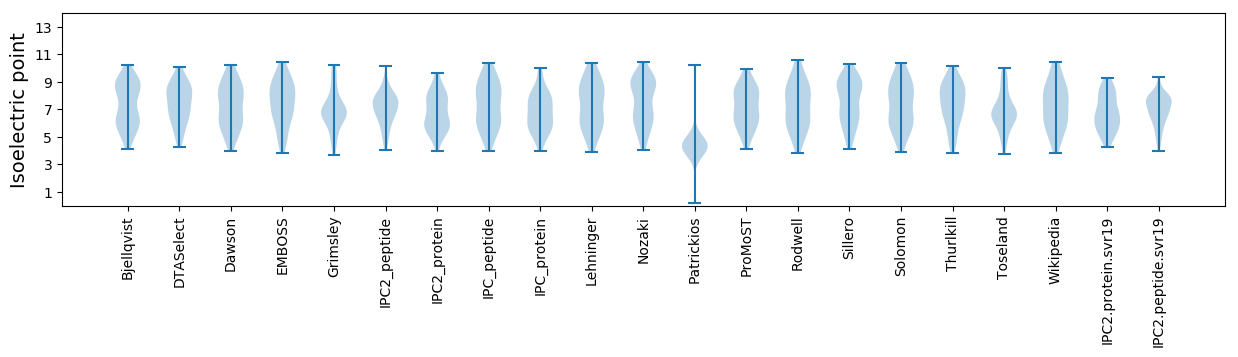
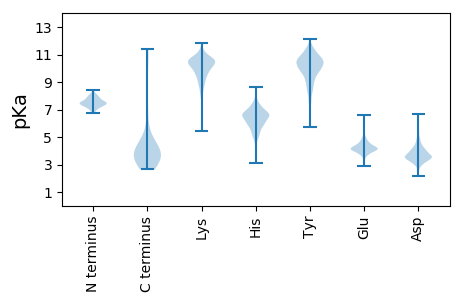
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q00146|VG36_ICHVA Uncharacterized protein ORF36 OS=Ictalurid herpesvirus 1 (strain Auburn) OX=766178 GN=ORF36 PE=4 SV=1
MM1 pKa = 7.28TPVPPSSLRR10 pKa = 11.84SIAPLSTVFLFTDD23 pKa = 3.87NVRR26 pKa = 11.84SNSWFVCKK34 pKa = 10.2IVYY37 pKa = 9.81RR38 pKa = 11.84CLSPRR43 pKa = 11.84RR44 pKa = 11.84IISPSICTAFSCWANHH60 pKa = 5.15GLRR63 pKa = 11.84LSSTAAMRR71 pKa = 11.84RR72 pKa = 11.84SSSSTFLFMASTRR85 pKa = 11.84MSSSTFVCFIFVFCVSRR102 pKa = 11.84TAMVFWSFSEE112 pKa = 4.35STSLLWCIISSSMRR126 pKa = 11.84LCFSITSGSTT136 pKa = 2.85
MM1 pKa = 7.28TPVPPSSLRR10 pKa = 11.84SIAPLSTVFLFTDD23 pKa = 3.87NVRR26 pKa = 11.84SNSWFVCKK34 pKa = 10.2IVYY37 pKa = 9.81RR38 pKa = 11.84CLSPRR43 pKa = 11.84RR44 pKa = 11.84IISPSICTAFSCWANHH60 pKa = 5.15GLRR63 pKa = 11.84LSSTAAMRR71 pKa = 11.84RR72 pKa = 11.84SSSSTFLFMASTRR85 pKa = 11.84MSSSTFVCFIFVFCVSRR102 pKa = 11.84TAMVFWSFSEE112 pKa = 4.35STSLLWCIISSSMRR126 pKa = 11.84LCFSITSGSTT136 pKa = 2.85
Molecular weight: 15.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
33924 |
82 |
1556 |
446.4 |
49.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.464 ± 0.271 | 1.816 ± 0.149 |
5.636 ± 0.158 | 5.769 ± 0.241 |
4.021 ± 0.182 | 7.116 ± 0.206 |
2.273 ± 0.09 | 5.545 ± 0.175 |
4.345 ± 0.256 | 9.114 ± 0.24 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.803 ± 0.112 | 3.44 ± 0.164 |
6.143 ± 0.321 | 2.411 ± 0.165 |
6.624 ± 0.241 | 6.367 ± 0.176 |
7.623 ± 0.498 | 7.502 ± 0.146 |
1.197 ± 0.087 | 2.792 ± 0.14 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
