
Salegentibacter mishustinae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Salegentibacter
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
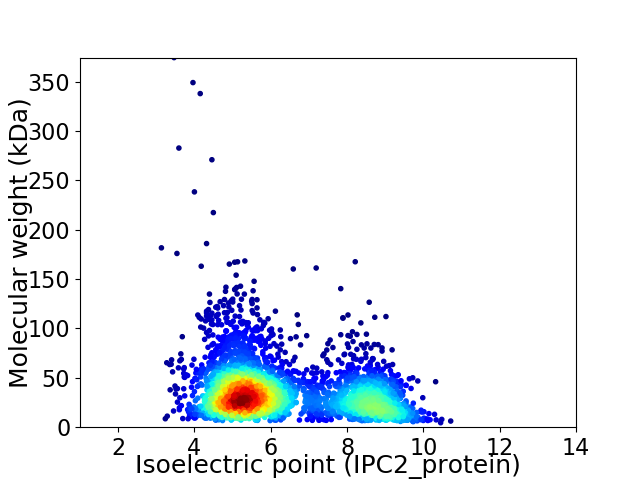
Virtual 2D-PAGE plot for 3284 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q9ZFK8|A0A0Q9ZFK8_9FLAO DNA-directed RNA polymerase subunit beta OS=Salegentibacter mishustinae OX=270918 GN=rpoB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 10.53NIFKK6 pKa = 10.35TSLAVLSLVFFSACSNDD23 pKa = 4.09DD24 pKa = 3.53YY25 pKa = 11.57DD26 pKa = 5.15IGDD29 pKa = 3.39ITTPSNLSVTAEE41 pKa = 3.92VVGQTEE47 pKa = 4.28EE48 pKa = 4.12MPNGDD53 pKa = 2.83GSGAVSFTASADD65 pKa = 3.26NAMTYY70 pKa = 10.58KK71 pKa = 10.77FIYY74 pKa = 9.96GDD76 pKa = 3.53GFEE79 pKa = 4.54EE80 pKa = 4.82VSASGEE86 pKa = 4.27TTHH89 pKa = 6.77SFNEE93 pKa = 4.17NGVNDD98 pKa = 3.7YY99 pKa = 9.27TVTVVASGTGGASSNMTTTVTVFSDD124 pKa = 4.34FSDD127 pKa = 4.4PEE129 pKa = 4.37TKK131 pKa = 10.25EE132 pKa = 3.91LLTGGSSKK140 pKa = 8.27TWYY143 pKa = 9.79VAASQPAHH151 pKa = 6.62LGVGPSSGEE160 pKa = 3.98GFTAPIYY167 pKa = 9.79YY168 pKa = 10.31AAAPFEE174 pKa = 4.25KK175 pKa = 10.44AGADD179 pKa = 3.5VSSCFYY185 pKa = 9.46TDD187 pKa = 3.64EE188 pKa = 4.28MTFSLNEE195 pKa = 3.76NDD197 pKa = 4.05NIVYY201 pKa = 10.39NYY203 pKa = 10.04NNNGLTFVNVAYY215 pKa = 9.53TGDD218 pKa = 3.51FGGDD222 pKa = 3.4GSEE225 pKa = 4.3DD226 pKa = 3.32QCLDD230 pKa = 3.49FGNTGDD236 pKa = 4.63FSASLSPSTSGLPEE250 pKa = 4.7DD251 pKa = 4.07ATTGTVINIAGGGTMSYY268 pKa = 10.9YY269 pKa = 10.62VGSSSYY275 pKa = 10.59EE276 pKa = 3.84VLNITPTTMDD286 pKa = 3.0VRR288 pKa = 11.84VIPGNDD294 pKa = 3.45PALAWYY300 pKa = 10.15LKK302 pKa = 9.38FTTSQGGEE310 pKa = 3.95EE311 pKa = 4.14EE312 pKa = 4.87EE313 pKa = 4.29EE314 pKa = 4.44FEE316 pKa = 4.76SQFNTEE322 pKa = 3.41IWSDD326 pKa = 3.78EE327 pKa = 4.03FDD329 pKa = 3.78GDD331 pKa = 5.66ALNTDD336 pKa = 2.71NWNYY340 pKa = 7.77EE341 pKa = 4.08TGNGSNGWGNNEE353 pKa = 3.45AQYY356 pKa = 9.69YY357 pKa = 9.02TDD359 pKa = 4.96AEE361 pKa = 4.77DD362 pKa = 3.59NVKK365 pKa = 10.2VEE367 pKa = 4.21NGNLVITAKK376 pKa = 10.54RR377 pKa = 11.84EE378 pKa = 4.12AEE380 pKa = 4.06SGFDD384 pKa = 3.41FTSARR389 pKa = 11.84ITTKK393 pKa = 10.92DD394 pKa = 2.85KK395 pKa = 11.51FEE397 pKa = 4.15FTFGRR402 pKa = 11.84VEE404 pKa = 4.06VRR406 pKa = 11.84AKK408 pKa = 10.65LPAGGGTWPAIWMLGAAFPEE428 pKa = 4.61EE429 pKa = 4.24SWPGVGEE436 pKa = 3.84MDD438 pKa = 3.18IMEE441 pKa = 4.59FVGNDD446 pKa = 3.34PDD448 pKa = 4.82RR449 pKa = 11.84ISSALHH455 pKa = 5.49FPGNSGGNAIVGDD468 pKa = 4.17TEE470 pKa = 4.34VSDD473 pKa = 3.91VTSEE477 pKa = 3.56FHH479 pKa = 6.9IYY481 pKa = 8.66EE482 pKa = 4.34VEE484 pKa = 3.76WTAEE488 pKa = 4.13KK489 pKa = 9.5ITFLMDD495 pKa = 3.67GVPHH499 pKa = 7.7LEE501 pKa = 3.84FDD503 pKa = 4.53NNDD506 pKa = 3.27STPFQEE512 pKa = 5.63DD513 pKa = 3.37FFIILNVAMGGTLGGNIADD532 pKa = 4.73DD533 pKa = 4.09FQEE536 pKa = 4.24SSMEE540 pKa = 3.64VDD542 pKa = 3.49YY543 pKa = 11.86VKK545 pKa = 10.74VFQEE549 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 10.53NIFKK6 pKa = 10.35TSLAVLSLVFFSACSNDD23 pKa = 4.09DD24 pKa = 3.53YY25 pKa = 11.57DD26 pKa = 5.15IGDD29 pKa = 3.39ITTPSNLSVTAEE41 pKa = 3.92VVGQTEE47 pKa = 4.28EE48 pKa = 4.12MPNGDD53 pKa = 2.83GSGAVSFTASADD65 pKa = 3.26NAMTYY70 pKa = 10.58KK71 pKa = 10.77FIYY74 pKa = 9.96GDD76 pKa = 3.53GFEE79 pKa = 4.54EE80 pKa = 4.82VSASGEE86 pKa = 4.27TTHH89 pKa = 6.77SFNEE93 pKa = 4.17NGVNDD98 pKa = 3.7YY99 pKa = 9.27TVTVVASGTGGASSNMTTTVTVFSDD124 pKa = 4.34FSDD127 pKa = 4.4PEE129 pKa = 4.37TKK131 pKa = 10.25EE132 pKa = 3.91LLTGGSSKK140 pKa = 8.27TWYY143 pKa = 9.79VAASQPAHH151 pKa = 6.62LGVGPSSGEE160 pKa = 3.98GFTAPIYY167 pKa = 9.79YY168 pKa = 10.31AAAPFEE174 pKa = 4.25KK175 pKa = 10.44AGADD179 pKa = 3.5VSSCFYY185 pKa = 9.46TDD187 pKa = 3.64EE188 pKa = 4.28MTFSLNEE195 pKa = 3.76NDD197 pKa = 4.05NIVYY201 pKa = 10.39NYY203 pKa = 10.04NNNGLTFVNVAYY215 pKa = 9.53TGDD218 pKa = 3.51FGGDD222 pKa = 3.4GSEE225 pKa = 4.3DD226 pKa = 3.32QCLDD230 pKa = 3.49FGNTGDD236 pKa = 4.63FSASLSPSTSGLPEE250 pKa = 4.7DD251 pKa = 4.07ATTGTVINIAGGGTMSYY268 pKa = 10.9YY269 pKa = 10.62VGSSSYY275 pKa = 10.59EE276 pKa = 3.84VLNITPTTMDD286 pKa = 3.0VRR288 pKa = 11.84VIPGNDD294 pKa = 3.45PALAWYY300 pKa = 10.15LKK302 pKa = 9.38FTTSQGGEE310 pKa = 3.95EE311 pKa = 4.14EE312 pKa = 4.87EE313 pKa = 4.29EE314 pKa = 4.44FEE316 pKa = 4.76SQFNTEE322 pKa = 3.41IWSDD326 pKa = 3.78EE327 pKa = 4.03FDD329 pKa = 3.78GDD331 pKa = 5.66ALNTDD336 pKa = 2.71NWNYY340 pKa = 7.77EE341 pKa = 4.08TGNGSNGWGNNEE353 pKa = 3.45AQYY356 pKa = 9.69YY357 pKa = 9.02TDD359 pKa = 4.96AEE361 pKa = 4.77DD362 pKa = 3.59NVKK365 pKa = 10.2VEE367 pKa = 4.21NGNLVITAKK376 pKa = 10.54RR377 pKa = 11.84EE378 pKa = 4.12AEE380 pKa = 4.06SGFDD384 pKa = 3.41FTSARR389 pKa = 11.84ITTKK393 pKa = 10.92DD394 pKa = 2.85KK395 pKa = 11.51FEE397 pKa = 4.15FTFGRR402 pKa = 11.84VEE404 pKa = 4.06VRR406 pKa = 11.84AKK408 pKa = 10.65LPAGGGTWPAIWMLGAAFPEE428 pKa = 4.61EE429 pKa = 4.24SWPGVGEE436 pKa = 3.84MDD438 pKa = 3.18IMEE441 pKa = 4.59FVGNDD446 pKa = 3.34PDD448 pKa = 4.82RR449 pKa = 11.84ISSALHH455 pKa = 5.49FPGNSGGNAIVGDD468 pKa = 4.17TEE470 pKa = 4.34VSDD473 pKa = 3.91VTSEE477 pKa = 3.56FHH479 pKa = 6.9IYY481 pKa = 8.66EE482 pKa = 4.34VEE484 pKa = 3.76WTAEE488 pKa = 4.13KK489 pKa = 9.5ITFLMDD495 pKa = 3.67GVPHH499 pKa = 7.7LEE501 pKa = 3.84FDD503 pKa = 4.53NNDD506 pKa = 3.27STPFQEE512 pKa = 5.63DD513 pKa = 3.37FFIILNVAMGGTLGGNIADD532 pKa = 4.73DD533 pKa = 4.09FQEE536 pKa = 4.24SSMEE540 pKa = 3.64VDD542 pKa = 3.49YY543 pKa = 11.86VKK545 pKa = 10.74VFQEE549 pKa = 3.96
Molecular weight: 59.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q9ZA26|A0A0Q9ZA26_9FLAO Phosphoribosylpyrophosphate synthetase OS=Salegentibacter mishustinae OX=270918 GN=APR42_15290 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.34KK41 pKa = 10.36ISVSSEE47 pKa = 3.42NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.34KK41 pKa = 10.36ISVSSEE47 pKa = 3.42NRR49 pKa = 11.84HH50 pKa = 4.47KK51 pKa = 10.5HH52 pKa = 4.49
Molecular weight: 6.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1108007 |
38 |
3537 |
337.4 |
38.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.469 ± 0.04 | 0.653 ± 0.015 |
5.538 ± 0.038 | 7.841 ± 0.043 |
5.285 ± 0.036 | 6.461 ± 0.042 |
1.712 ± 0.021 | 7.669 ± 0.037 |
7.764 ± 0.057 | 9.371 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.208 ± 0.026 | 5.919 ± 0.046 |
3.48 ± 0.026 | 3.426 ± 0.025 |
3.708 ± 0.03 | 6.364 ± 0.029 |
5.217 ± 0.041 | 5.921 ± 0.032 |
1.056 ± 0.015 | 3.937 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
