
Marinimicrobium koreense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Cellvibrionaceae; Marinimicrobium
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
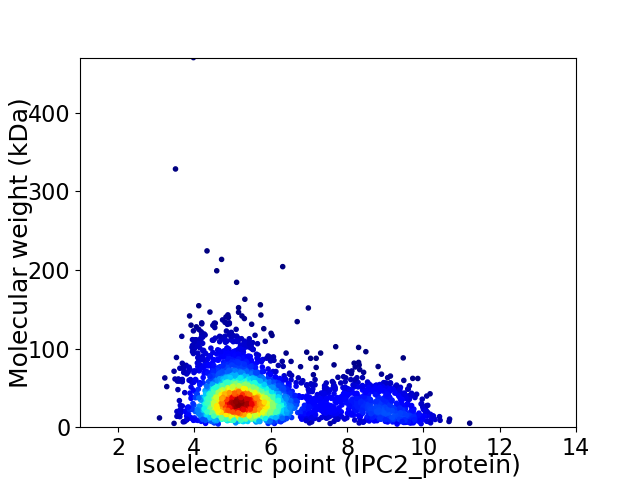
Virtual 2D-PAGE plot for 3203 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N1P249|A0A3N1P249_9GAMM Flavin-dependent dehydrogenase OS=Marinimicrobium koreense OX=306545 GN=EDC38_2304 PE=4 SV=1
MM1 pKa = 7.7KK2 pKa = 10.19YY3 pKa = 9.57IASAVLAAALLAPAYY18 pKa = 9.62GAVAAAPSNLALDD31 pKa = 4.41KK32 pKa = 10.48PVSASSSQNRR42 pKa = 11.84NRR44 pKa = 11.84DD45 pKa = 2.85AEE47 pKa = 4.12FAVDD51 pKa = 3.86GNVGTRR57 pKa = 11.84WSSDD61 pKa = 3.11FSDD64 pKa = 4.23GNWIIVDD71 pKa = 4.32LEE73 pKa = 4.23SVYY76 pKa = 10.82DD77 pKa = 3.64ISSVILQWEE86 pKa = 4.04TAYY89 pKa = 10.05PEE91 pKa = 4.14QYY93 pKa = 10.61NLEE96 pKa = 4.1VSLDD100 pKa = 3.54GSNWEE105 pKa = 4.18TVSAVTSSNGGVDD118 pKa = 3.49SFAMTRR124 pKa = 11.84SARR127 pKa = 11.84YY128 pKa = 9.73VRR130 pKa = 11.84VEE132 pKa = 3.94SVKK135 pKa = 10.57RR136 pKa = 11.84ATKK139 pKa = 9.52WGVSLWEE146 pKa = 3.95LEE148 pKa = 4.54VYY150 pKa = 10.22GVEE153 pKa = 4.37PEE155 pKa = 4.52PEE157 pKa = 4.32PEE159 pKa = 4.35PEE161 pKa = 4.37PEE163 pKa = 4.45PEE165 pKa = 4.62PEE167 pKa = 4.25TEE169 pKa = 4.35PEE171 pKa = 4.12STTDD175 pKa = 3.67SEE177 pKa = 4.77TTSGDD182 pKa = 3.52GANLALNQTMFASSSEE198 pKa = 4.04NSSRR202 pKa = 11.84LPEE205 pKa = 4.04EE206 pKa = 4.97AVDD209 pKa = 3.68GDD211 pKa = 4.26MNSRR215 pKa = 11.84WASEE219 pKa = 3.98YY220 pKa = 11.51GEE222 pKa = 4.48DD223 pKa = 3.34QWIAVDD229 pKa = 5.43LGATYY234 pKa = 10.88DD235 pKa = 3.82LSSLILRR242 pKa = 11.84WEE244 pKa = 4.08VAHH247 pKa = 7.22AEE249 pKa = 4.24DD250 pKa = 5.01YY251 pKa = 10.89DD252 pKa = 3.99IQVSADD258 pKa = 3.35ANDD261 pKa = 3.42WQTAQSVTNSDD272 pKa = 3.01GGEE275 pKa = 4.03DD276 pKa = 3.8VVEE279 pKa = 4.67LSATGRR285 pKa = 11.84YY286 pKa = 7.77VRR288 pKa = 11.84IQSVRR293 pKa = 11.84RR294 pKa = 11.84ATQWGISLWEE304 pKa = 4.01LEE306 pKa = 4.59VYY308 pKa = 10.2GAEE311 pKa = 4.34PYY313 pKa = 9.86VAPEE317 pKa = 3.9PQLVSEE323 pKa = 4.92GRR325 pKa = 11.84PATASSTEE333 pKa = 3.79RR334 pKa = 11.84DD335 pKa = 3.21AYY337 pKa = 10.72APEE340 pKa = 3.89NAFDD344 pKa = 4.32GSGSTRR350 pKa = 11.84WASVYY355 pKa = 11.07GDD357 pKa = 3.94TAWLQVDD364 pKa = 4.56LGEE367 pKa = 4.21AHH369 pKa = 6.92QITGINLDD377 pKa = 3.23WEE379 pKa = 4.51TAYY382 pKa = 10.95SSEE385 pKa = 4.03YY386 pKa = 10.25QIQTSYY392 pKa = 11.5DD393 pKa = 3.58GQSWTTVEE401 pKa = 5.16HH402 pKa = 6.61IVDD405 pKa = 4.02GNGGTDD411 pKa = 3.32EE412 pKa = 4.85LVVDD416 pKa = 3.26GHH418 pKa = 5.56GRR420 pKa = 11.84YY421 pKa = 9.29VRR423 pKa = 11.84MMGLARR429 pKa = 11.84ATSWGHH435 pKa = 5.82SLWEE439 pKa = 4.07MQVFGYY445 pKa = 9.52PADD448 pKa = 4.18SADD451 pKa = 3.89SPSDD455 pKa = 3.51GTDD458 pKa = 2.7GSTDD462 pKa = 3.6DD463 pKa = 4.07GTGSEE468 pKa = 4.72TDD470 pKa = 3.35TGSDD474 pKa = 3.38TDD476 pKa = 3.64TGSDD480 pKa = 4.18DD481 pKa = 3.92GSTGDD486 pKa = 3.5GSTGGEE492 pKa = 3.83EE493 pKa = 4.13STEE496 pKa = 4.05EE497 pKa = 4.03EE498 pKa = 4.68TVDD501 pKa = 3.37TTAPSAPGSLNDD513 pKa = 3.78TAVGTSYY520 pKa = 9.48VTLSWSEE527 pKa = 4.05STDD530 pKa = 3.24NEE532 pKa = 3.91AVMSYY537 pKa = 9.57EE538 pKa = 4.08VYY540 pKa = 10.51RR541 pKa = 11.84DD542 pKa = 3.35GARR545 pKa = 11.84ITEE548 pKa = 4.37LLAPEE553 pKa = 4.48TSYY556 pKa = 11.7TDD558 pKa = 3.17TGLSSGTTYY567 pKa = 10.45QYY569 pKa = 11.18SVRR572 pKa = 11.84AGDD575 pKa = 3.97AAGNWSEE582 pKa = 3.98NSTTLVVTTNSEE594 pKa = 4.12QVVGDD599 pKa = 3.79GVVLEE604 pKa = 4.63WTTPDD609 pKa = 3.05QRR611 pKa = 11.84EE612 pKa = 3.99NGSYY616 pKa = 11.1LEE618 pKa = 4.15LNEE621 pKa = 3.8IGGYY625 pKa = 7.67EE626 pKa = 3.78IRR628 pKa = 11.84YY629 pKa = 7.61QQGADD634 pKa = 3.56GEE636 pKa = 4.71SQSVVVNDD644 pKa = 4.9AYY646 pKa = 10.42ATSHH650 pKa = 6.05EE651 pKa = 4.82LNLEE655 pKa = 4.11DD656 pKa = 5.56GIYY659 pKa = 9.79EE660 pKa = 3.75FSIAVFDD667 pKa = 4.17VNGLYY672 pKa = 10.53SQFVPIDD679 pKa = 3.64PVPP682 pKa = 3.5
MM1 pKa = 7.7KK2 pKa = 10.19YY3 pKa = 9.57IASAVLAAALLAPAYY18 pKa = 9.62GAVAAAPSNLALDD31 pKa = 4.41KK32 pKa = 10.48PVSASSSQNRR42 pKa = 11.84NRR44 pKa = 11.84DD45 pKa = 2.85AEE47 pKa = 4.12FAVDD51 pKa = 3.86GNVGTRR57 pKa = 11.84WSSDD61 pKa = 3.11FSDD64 pKa = 4.23GNWIIVDD71 pKa = 4.32LEE73 pKa = 4.23SVYY76 pKa = 10.82DD77 pKa = 3.64ISSVILQWEE86 pKa = 4.04TAYY89 pKa = 10.05PEE91 pKa = 4.14QYY93 pKa = 10.61NLEE96 pKa = 4.1VSLDD100 pKa = 3.54GSNWEE105 pKa = 4.18TVSAVTSSNGGVDD118 pKa = 3.49SFAMTRR124 pKa = 11.84SARR127 pKa = 11.84YY128 pKa = 9.73VRR130 pKa = 11.84VEE132 pKa = 3.94SVKK135 pKa = 10.57RR136 pKa = 11.84ATKK139 pKa = 9.52WGVSLWEE146 pKa = 3.95LEE148 pKa = 4.54VYY150 pKa = 10.22GVEE153 pKa = 4.37PEE155 pKa = 4.52PEE157 pKa = 4.32PEE159 pKa = 4.35PEE161 pKa = 4.37PEE163 pKa = 4.45PEE165 pKa = 4.62PEE167 pKa = 4.25TEE169 pKa = 4.35PEE171 pKa = 4.12STTDD175 pKa = 3.67SEE177 pKa = 4.77TTSGDD182 pKa = 3.52GANLALNQTMFASSSEE198 pKa = 4.04NSSRR202 pKa = 11.84LPEE205 pKa = 4.04EE206 pKa = 4.97AVDD209 pKa = 3.68GDD211 pKa = 4.26MNSRR215 pKa = 11.84WASEE219 pKa = 3.98YY220 pKa = 11.51GEE222 pKa = 4.48DD223 pKa = 3.34QWIAVDD229 pKa = 5.43LGATYY234 pKa = 10.88DD235 pKa = 3.82LSSLILRR242 pKa = 11.84WEE244 pKa = 4.08VAHH247 pKa = 7.22AEE249 pKa = 4.24DD250 pKa = 5.01YY251 pKa = 10.89DD252 pKa = 3.99IQVSADD258 pKa = 3.35ANDD261 pKa = 3.42WQTAQSVTNSDD272 pKa = 3.01GGEE275 pKa = 4.03DD276 pKa = 3.8VVEE279 pKa = 4.67LSATGRR285 pKa = 11.84YY286 pKa = 7.77VRR288 pKa = 11.84IQSVRR293 pKa = 11.84RR294 pKa = 11.84ATQWGISLWEE304 pKa = 4.01LEE306 pKa = 4.59VYY308 pKa = 10.2GAEE311 pKa = 4.34PYY313 pKa = 9.86VAPEE317 pKa = 3.9PQLVSEE323 pKa = 4.92GRR325 pKa = 11.84PATASSTEE333 pKa = 3.79RR334 pKa = 11.84DD335 pKa = 3.21AYY337 pKa = 10.72APEE340 pKa = 3.89NAFDD344 pKa = 4.32GSGSTRR350 pKa = 11.84WASVYY355 pKa = 11.07GDD357 pKa = 3.94TAWLQVDD364 pKa = 4.56LGEE367 pKa = 4.21AHH369 pKa = 6.92QITGINLDD377 pKa = 3.23WEE379 pKa = 4.51TAYY382 pKa = 10.95SSEE385 pKa = 4.03YY386 pKa = 10.25QIQTSYY392 pKa = 11.5DD393 pKa = 3.58GQSWTTVEE401 pKa = 5.16HH402 pKa = 6.61IVDD405 pKa = 4.02GNGGTDD411 pKa = 3.32EE412 pKa = 4.85LVVDD416 pKa = 3.26GHH418 pKa = 5.56GRR420 pKa = 11.84YY421 pKa = 9.29VRR423 pKa = 11.84MMGLARR429 pKa = 11.84ATSWGHH435 pKa = 5.82SLWEE439 pKa = 4.07MQVFGYY445 pKa = 9.52PADD448 pKa = 4.18SADD451 pKa = 3.89SPSDD455 pKa = 3.51GTDD458 pKa = 2.7GSTDD462 pKa = 3.6DD463 pKa = 4.07GTGSEE468 pKa = 4.72TDD470 pKa = 3.35TGSDD474 pKa = 3.38TDD476 pKa = 3.64TGSDD480 pKa = 4.18DD481 pKa = 3.92GSTGDD486 pKa = 3.5GSTGGEE492 pKa = 3.83EE493 pKa = 4.13STEE496 pKa = 4.05EE497 pKa = 4.03EE498 pKa = 4.68TVDD501 pKa = 3.37TTAPSAPGSLNDD513 pKa = 3.78TAVGTSYY520 pKa = 9.48VTLSWSEE527 pKa = 4.05STDD530 pKa = 3.24NEE532 pKa = 3.91AVMSYY537 pKa = 9.57EE538 pKa = 4.08VYY540 pKa = 10.51RR541 pKa = 11.84DD542 pKa = 3.35GARR545 pKa = 11.84ITEE548 pKa = 4.37LLAPEE553 pKa = 4.48TSYY556 pKa = 11.7TDD558 pKa = 3.17TGLSSGTTYY567 pKa = 10.45QYY569 pKa = 11.18SVRR572 pKa = 11.84AGDD575 pKa = 3.97AAGNWSEE582 pKa = 3.98NSTTLVVTTNSEE594 pKa = 4.12QVVGDD599 pKa = 3.79GVVLEE604 pKa = 4.63WTTPDD609 pKa = 3.05QRR611 pKa = 11.84EE612 pKa = 3.99NGSYY616 pKa = 11.1LEE618 pKa = 4.15LNEE621 pKa = 3.8IGGYY625 pKa = 7.67EE626 pKa = 3.78IRR628 pKa = 11.84YY629 pKa = 7.61QQGADD634 pKa = 3.56GEE636 pKa = 4.71SQSVVVNDD644 pKa = 4.9AYY646 pKa = 10.42ATSHH650 pKa = 6.05EE651 pKa = 4.82LNLEE655 pKa = 4.11DD656 pKa = 5.56GIYY659 pKa = 9.79EE660 pKa = 3.75FSIAVFDD667 pKa = 4.17VNGLYY672 pKa = 10.53SQFVPIDD679 pKa = 3.64PVPP682 pKa = 3.5
Molecular weight: 73.5 kDa
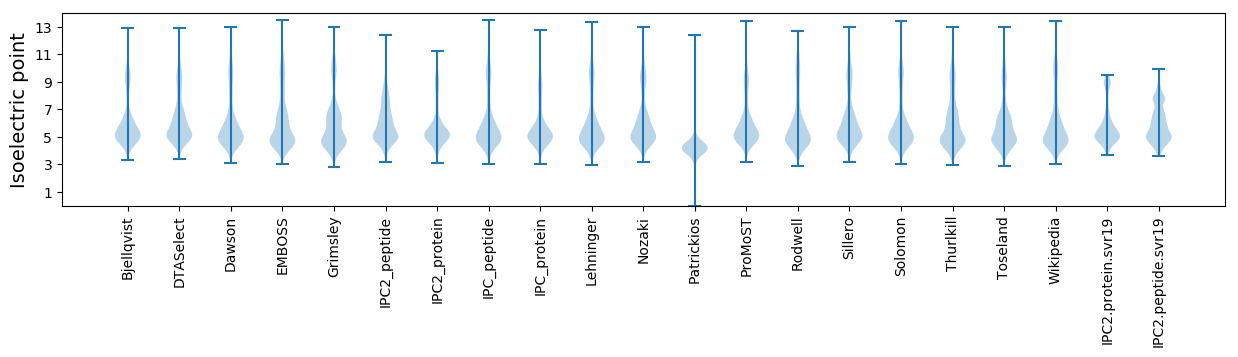
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N1P4B2|A0A3N1P4B2_9GAMM Carbohydrate binding protein with CBM6 domain OS=Marinimicrobium koreense OX=306545 GN=EDC38_0261 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84KK40 pKa = 8.7ALAVV44 pKa = 3.49
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.04GRR39 pKa = 11.84KK40 pKa = 8.7ALAVV44 pKa = 3.49
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1151955 |
34 |
4537 |
359.6 |
39.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.665 ± 0.048 | 0.877 ± 0.013 |
6.172 ± 0.037 | 6.781 ± 0.037 |
3.688 ± 0.029 | 7.737 ± 0.045 |
2.295 ± 0.025 | 4.797 ± 0.029 |
3.277 ± 0.036 | 10.644 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.263 ± 0.021 | 3.344 ± 0.031 |
4.802 ± 0.03 | 4.357 ± 0.03 |
6.505 ± 0.045 | 6.033 ± 0.04 |
5.239 ± 0.035 | 7.067 ± 0.036 |
1.55 ± 0.022 | 2.907 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
