
Arthrobacter sp. YC-RL1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter; unclassified Arthrobacter
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
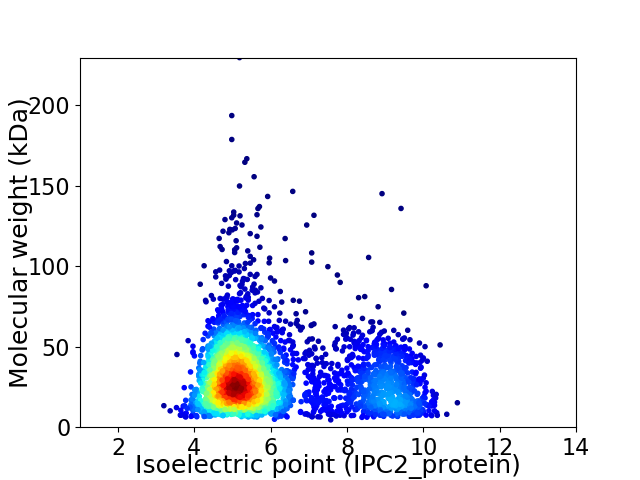
Virtual 2D-PAGE plot for 3439 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
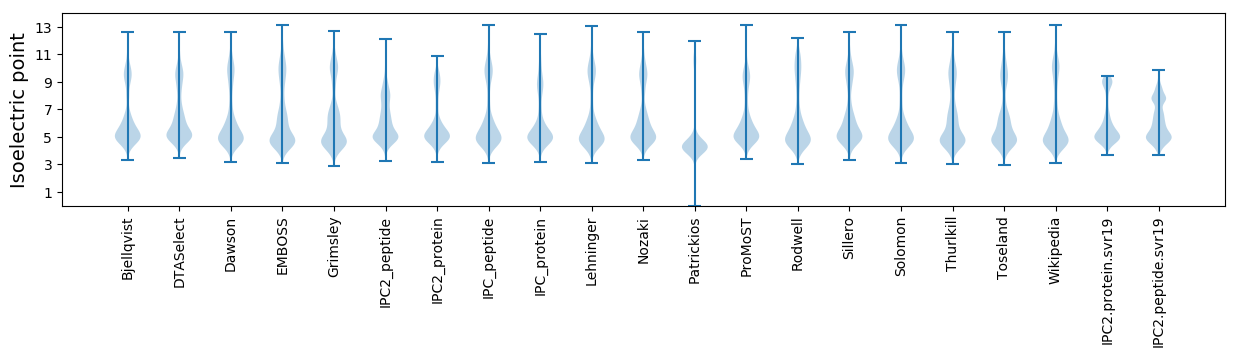
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H0ZTS0|A0A0H0ZTS0_9MICC Phosphomannomutase OS=Arthrobacter sp. YC-RL1 OX=1652545 GN=AA310_04950 PE=3 SV=1
MM1 pKa = 7.04YY2 pKa = 10.23RR3 pKa = 11.84LSATAAALALGLTACAPTSGEE24 pKa = 4.13DD25 pKa = 3.95GEE27 pKa = 5.62GNAADD32 pKa = 3.66ATKK35 pKa = 10.52TEE37 pKa = 4.35VPALQQYY44 pKa = 7.4YY45 pKa = 7.6TQQVDD50 pKa = 3.25WEE52 pKa = 4.66KK53 pKa = 10.98CGSTIEE59 pKa = 4.33CADD62 pKa = 3.28IEE64 pKa = 4.71VPLDD68 pKa = 3.3YY69 pKa = 10.77AAPEE73 pKa = 4.24KK74 pKa = 10.82DD75 pKa = 4.06SIMLALNRR83 pKa = 11.84RR84 pKa = 11.84AVDD87 pKa = 4.0GAQGNLLVNPGGPGGSGLDD106 pKa = 3.36MVAEE110 pKa = 4.64SVQLMLSNDD119 pKa = 3.32LQRR122 pKa = 11.84AYY124 pKa = 10.94NIIGFDD130 pKa = 3.55PRR132 pKa = 11.84GVGQSTPVTCQTDD145 pKa = 3.62KK146 pKa = 11.34EE147 pKa = 4.15IDD149 pKa = 3.18ASRR152 pKa = 11.84QEE154 pKa = 4.31NLKK157 pKa = 10.64AWDD160 pKa = 3.62PADD163 pKa = 4.3RR164 pKa = 11.84DD165 pKa = 3.65EE166 pKa = 5.27VEE168 pKa = 5.55ADD170 pKa = 3.34TEE172 pKa = 5.23DD173 pKa = 3.87YY174 pKa = 11.5AKK176 pKa = 10.87DD177 pKa = 3.5CAEE180 pKa = 4.04NTGDD184 pKa = 4.78LLGHH188 pKa = 6.49VDD190 pKa = 3.68TVSAAKK196 pKa = 10.85DD197 pKa = 3.23MDD199 pKa = 4.35IIRR202 pKa = 11.84AVLGDD207 pKa = 3.63TQLDD211 pKa = 3.92YY212 pKa = 11.55LGYY215 pKa = 10.5SYY217 pKa = 10.66GTFLGSTYY225 pKa = 11.39ADD227 pKa = 4.32LFPQKK232 pKa = 10.25VGRR235 pKa = 11.84FVLDD239 pKa = 3.52GAMDD243 pKa = 4.77PLATAQQMTLAQAQGFEE260 pKa = 4.34GEE262 pKa = 3.71IDD264 pKa = 3.4AWLQNCLEE272 pKa = 4.5GEE274 pKa = 4.21NCPFTGNLDD283 pKa = 3.59EE284 pKa = 6.57AKK286 pKa = 9.12TQLQDD291 pKa = 3.04FFARR295 pKa = 11.84IEE297 pKa = 4.06EE298 pKa = 4.21QPLEE302 pKa = 4.32SSDD305 pKa = 3.16GRR307 pKa = 11.84TVPIIDD313 pKa = 5.43FINGFIIPLYY323 pKa = 10.99DD324 pKa = 4.12DD325 pKa = 4.32MNWPYY330 pKa = 9.76LTQGMAAAMQGDD342 pKa = 4.33VDD344 pKa = 4.12QILYY348 pKa = 10.61FSDD351 pKa = 4.25LSADD355 pKa = 3.9RR356 pKa = 11.84QEE358 pKa = 4.58DD359 pKa = 3.48GSYY362 pKa = 11.23GSNSTDD368 pKa = 2.62AFTAVNCLDD377 pKa = 3.65RR378 pKa = 11.84PMDD381 pKa = 4.01ASEE384 pKa = 3.96QAMEE388 pKa = 4.56ADD390 pKa = 3.36AKK392 pKa = 10.86ALEE395 pKa = 4.49EE396 pKa = 4.14ASPTIGKK403 pKa = 9.11YY404 pKa = 9.66LAYY407 pKa = 10.5GEE409 pKa = 4.39LTCDD413 pKa = 2.57KK414 pKa = 9.76WSYY417 pKa = 10.5KK418 pKa = 9.91ATGQPEE424 pKa = 4.37SLDD427 pKa = 3.66APGSNQILVVGTTGDD442 pKa = 3.59PATPYY447 pKa = 10.75AWSQSLAEE455 pKa = 4.06QLQNATLLTYY465 pKa = 10.06EE466 pKa = 4.37GHH468 pKa = 6.0GHH470 pKa = 4.9TAYY473 pKa = 10.3GRR475 pKa = 11.84SNQCITDD482 pKa = 3.41AVDD485 pKa = 4.58GYY487 pKa = 11.15LIDD490 pKa = 4.31GKK492 pKa = 10.83VPEE495 pKa = 5.32AGTVCC500 pKa = 4.44
MM1 pKa = 7.04YY2 pKa = 10.23RR3 pKa = 11.84LSATAAALALGLTACAPTSGEE24 pKa = 4.13DD25 pKa = 3.95GEE27 pKa = 5.62GNAADD32 pKa = 3.66ATKK35 pKa = 10.52TEE37 pKa = 4.35VPALQQYY44 pKa = 7.4YY45 pKa = 7.6TQQVDD50 pKa = 3.25WEE52 pKa = 4.66KK53 pKa = 10.98CGSTIEE59 pKa = 4.33CADD62 pKa = 3.28IEE64 pKa = 4.71VPLDD68 pKa = 3.3YY69 pKa = 10.77AAPEE73 pKa = 4.24KK74 pKa = 10.82DD75 pKa = 4.06SIMLALNRR83 pKa = 11.84RR84 pKa = 11.84AVDD87 pKa = 4.0GAQGNLLVNPGGPGGSGLDD106 pKa = 3.36MVAEE110 pKa = 4.64SVQLMLSNDD119 pKa = 3.32LQRR122 pKa = 11.84AYY124 pKa = 10.94NIIGFDD130 pKa = 3.55PRR132 pKa = 11.84GVGQSTPVTCQTDD145 pKa = 3.62KK146 pKa = 11.34EE147 pKa = 4.15IDD149 pKa = 3.18ASRR152 pKa = 11.84QEE154 pKa = 4.31NLKK157 pKa = 10.64AWDD160 pKa = 3.62PADD163 pKa = 4.3RR164 pKa = 11.84DD165 pKa = 3.65EE166 pKa = 5.27VEE168 pKa = 5.55ADD170 pKa = 3.34TEE172 pKa = 5.23DD173 pKa = 3.87YY174 pKa = 11.5AKK176 pKa = 10.87DD177 pKa = 3.5CAEE180 pKa = 4.04NTGDD184 pKa = 4.78LLGHH188 pKa = 6.49VDD190 pKa = 3.68TVSAAKK196 pKa = 10.85DD197 pKa = 3.23MDD199 pKa = 4.35IIRR202 pKa = 11.84AVLGDD207 pKa = 3.63TQLDD211 pKa = 3.92YY212 pKa = 11.55LGYY215 pKa = 10.5SYY217 pKa = 10.66GTFLGSTYY225 pKa = 11.39ADD227 pKa = 4.32LFPQKK232 pKa = 10.25VGRR235 pKa = 11.84FVLDD239 pKa = 3.52GAMDD243 pKa = 4.77PLATAQQMTLAQAQGFEE260 pKa = 4.34GEE262 pKa = 3.71IDD264 pKa = 3.4AWLQNCLEE272 pKa = 4.5GEE274 pKa = 4.21NCPFTGNLDD283 pKa = 3.59EE284 pKa = 6.57AKK286 pKa = 9.12TQLQDD291 pKa = 3.04FFARR295 pKa = 11.84IEE297 pKa = 4.06EE298 pKa = 4.21QPLEE302 pKa = 4.32SSDD305 pKa = 3.16GRR307 pKa = 11.84TVPIIDD313 pKa = 5.43FINGFIIPLYY323 pKa = 10.99DD324 pKa = 4.12DD325 pKa = 4.32MNWPYY330 pKa = 9.76LTQGMAAAMQGDD342 pKa = 4.33VDD344 pKa = 4.12QILYY348 pKa = 10.61FSDD351 pKa = 4.25LSADD355 pKa = 3.9RR356 pKa = 11.84QEE358 pKa = 4.58DD359 pKa = 3.48GSYY362 pKa = 11.23GSNSTDD368 pKa = 2.62AFTAVNCLDD377 pKa = 3.65RR378 pKa = 11.84PMDD381 pKa = 4.01ASEE384 pKa = 3.96QAMEE388 pKa = 4.56ADD390 pKa = 3.36AKK392 pKa = 10.86ALEE395 pKa = 4.49EE396 pKa = 4.14ASPTIGKK403 pKa = 9.11YY404 pKa = 9.66LAYY407 pKa = 10.5GEE409 pKa = 4.39LTCDD413 pKa = 2.57KK414 pKa = 9.76WSYY417 pKa = 10.5KK418 pKa = 9.91ATGQPEE424 pKa = 4.37SLDD427 pKa = 3.66APGSNQILVVGTTGDD442 pKa = 3.59PATPYY447 pKa = 10.75AWSQSLAEE455 pKa = 4.06QLQNATLLTYY465 pKa = 10.06EE466 pKa = 4.37GHH468 pKa = 6.0GHH470 pKa = 4.9TAYY473 pKa = 10.3GRR475 pKa = 11.84SNQCITDD482 pKa = 3.41AVDD485 pKa = 4.58GYY487 pKa = 11.15LIDD490 pKa = 4.31GKK492 pKa = 10.83VPEE495 pKa = 5.32AGTVCC500 pKa = 4.44
Molecular weight: 53.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H0ZND4|A0A0H0ZND4_9MICC TfoX_N domain-containing protein OS=Arthrobacter sp. YC-RL1 OX=1652545 GN=AA310_09315 PE=4 SV=1
MM1 pKa = 7.42DD2 pKa = 3.12QQMRR6 pKa = 11.84RR7 pKa = 11.84NPTRR11 pKa = 11.84TRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.12NRR17 pKa = 11.84PNRR20 pKa = 11.84QRR22 pKa = 11.84ATTQQRR28 pKa = 11.84RR29 pKa = 11.84QQRR32 pKa = 11.84NQKK35 pKa = 9.18RR36 pKa = 11.84RR37 pKa = 11.84QLSTTQPRR45 pKa = 11.84RR46 pKa = 11.84NAVEE50 pKa = 3.84NQPAKK55 pKa = 9.58HH56 pKa = 5.91HH57 pKa = 6.11PNRR60 pKa = 11.84RR61 pKa = 11.84TLRR64 pKa = 11.84QRR66 pKa = 11.84PTRR69 pKa = 11.84SRR71 pKa = 11.84HH72 pKa = 5.65RR73 pKa = 11.84LDD75 pKa = 3.36NPKK78 pKa = 10.09LVHH81 pKa = 6.24QSRR84 pKa = 11.84TTHH87 pKa = 4.36QRR89 pKa = 11.84TRR91 pKa = 11.84NRR93 pKa = 11.84THH95 pKa = 7.41RR96 pKa = 11.84PPSQPRR102 pKa = 11.84KK103 pKa = 10.07DD104 pKa = 3.45RR105 pKa = 11.84TDD107 pKa = 3.35RR108 pKa = 11.84ATNAHH113 pKa = 6.59ASTRR117 pKa = 11.84ALATRR122 pKa = 11.84KK123 pKa = 9.56RR124 pKa = 3.68
MM1 pKa = 7.42DD2 pKa = 3.12QQMRR6 pKa = 11.84RR7 pKa = 11.84NPTRR11 pKa = 11.84TRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.12NRR17 pKa = 11.84PNRR20 pKa = 11.84QRR22 pKa = 11.84ATTQQRR28 pKa = 11.84RR29 pKa = 11.84QQRR32 pKa = 11.84NQKK35 pKa = 9.18RR36 pKa = 11.84RR37 pKa = 11.84QLSTTQPRR45 pKa = 11.84RR46 pKa = 11.84NAVEE50 pKa = 3.84NQPAKK55 pKa = 9.58HH56 pKa = 5.91HH57 pKa = 6.11PNRR60 pKa = 11.84RR61 pKa = 11.84TLRR64 pKa = 11.84QRR66 pKa = 11.84PTRR69 pKa = 11.84SRR71 pKa = 11.84HH72 pKa = 5.65RR73 pKa = 11.84LDD75 pKa = 3.36NPKK78 pKa = 10.09LVHH81 pKa = 6.24QSRR84 pKa = 11.84TTHH87 pKa = 4.36QRR89 pKa = 11.84TRR91 pKa = 11.84NRR93 pKa = 11.84THH95 pKa = 7.41RR96 pKa = 11.84PPSQPRR102 pKa = 11.84KK103 pKa = 10.07DD104 pKa = 3.45RR105 pKa = 11.84TDD107 pKa = 3.35RR108 pKa = 11.84ATNAHH113 pKa = 6.59ASTRR117 pKa = 11.84ALATRR122 pKa = 11.84KK123 pKa = 9.56RR124 pKa = 3.68
Molecular weight: 15.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1069183 |
41 |
2163 |
310.9 |
33.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.529 ± 0.06 | 0.64 ± 0.011 |
5.454 ± 0.037 | 5.995 ± 0.045 |
3.332 ± 0.029 | 8.545 ± 0.04 |
2.114 ± 0.021 | 4.867 ± 0.029 |
3.157 ± 0.032 | 10.526 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.262 ± 0.018 | 2.681 ± 0.017 |
4.858 ± 0.027 | 3.669 ± 0.024 |
6.137 ± 0.038 | 6.107 ± 0.029 |
5.7 ± 0.037 | 7.872 ± 0.038 |
1.391 ± 0.016 | 2.162 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
