
Egaro virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
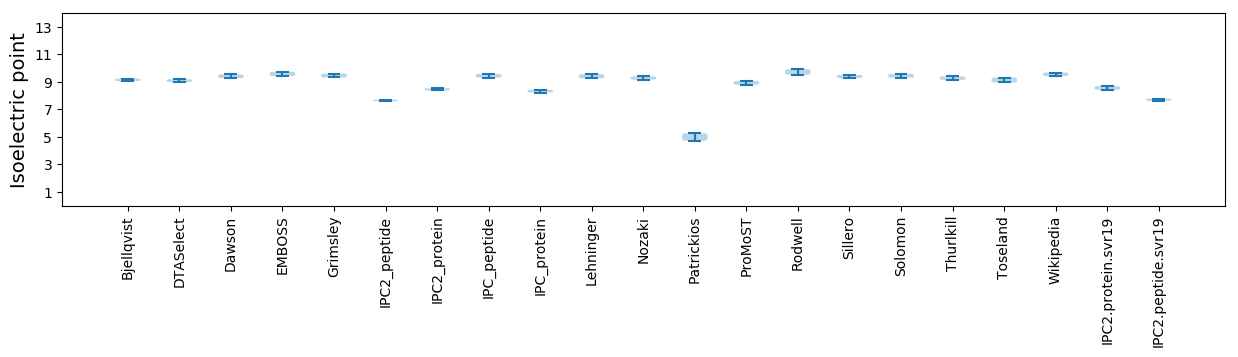
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1B2RVN9|A0A1B2RVN9_9VIRU RNA-dependent RNA polymerase OS=Egaro virus OX=1888318 PE=4 SV=1
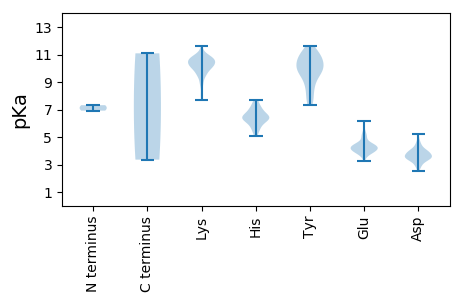
MM1 pKa = 7.32MNMRR5 pKa = 11.84LNPSSEE11 pKa = 4.02PLDD14 pKa = 3.81VQQLDD19 pKa = 3.95FLTEE23 pKa = 4.04LSPFPVFNLQNAQKK37 pKa = 9.01MQIHH41 pKa = 6.37PPKK44 pKa = 10.86VNMTGGFVSVARR56 pKa = 11.84SFGKK60 pKa = 9.87EE61 pKa = 3.29AARR64 pKa = 11.84DD65 pKa = 3.53RR66 pKa = 11.84RR67 pKa = 11.84LLGGALAYY75 pKa = 7.6FTQNEE80 pKa = 4.23VAEE83 pKa = 4.37ILQKK87 pKa = 9.64TVFCYY92 pKa = 9.35GTAKK96 pKa = 10.67GFLTRR101 pKa = 11.84LKK103 pKa = 10.59EE104 pKa = 3.92LTTRR108 pKa = 11.84DD109 pKa = 3.62CKK111 pKa = 10.48TPYY114 pKa = 10.25SISQTVSILNKK125 pKa = 9.95RR126 pKa = 11.84MPVLAEE132 pKa = 4.4EE133 pKa = 4.46LPSWEE138 pKa = 4.9DD139 pKa = 3.38FLSLLEE145 pKa = 4.37NTDD148 pKa = 3.38LTYY151 pKa = 10.67SASAGPPYY159 pKa = 9.71WRR161 pKa = 11.84SKK163 pKa = 10.95LMSLEE168 pKa = 4.09DD169 pKa = 3.68MEE171 pKa = 6.18LEE173 pKa = 4.13VLPLVLEE180 pKa = 4.17HH181 pKa = 6.36MKK183 pKa = 10.82AGTLNQLYY191 pKa = 9.64HH192 pKa = 6.29LHH194 pKa = 7.14PEE196 pKa = 4.29LFLVEE201 pKa = 4.67IKK203 pKa = 10.98NKK205 pKa = 9.72LDD207 pKa = 3.42RR208 pKa = 11.84YY209 pKa = 7.93EE210 pKa = 3.95TAKK213 pKa = 10.61FDD215 pKa = 4.16VKK217 pKa = 10.47CRR219 pKa = 11.84PYY221 pKa = 11.27GNVPFHH227 pKa = 6.41MGFLASMIVQPFCKK241 pKa = 10.01KK242 pKa = 10.4LKK244 pKa = 9.58LWHH247 pKa = 6.9NSDD250 pKa = 2.71SWNAYY255 pKa = 8.48GFSFAEE261 pKa = 4.41GSYY264 pKa = 10.95LEE266 pKa = 4.52LYY268 pKa = 9.91EE269 pKa = 4.2RR270 pKa = 11.84LKK272 pKa = 10.7KK273 pKa = 10.58FKK275 pKa = 10.71RR276 pKa = 11.84GDD278 pKa = 3.45EE279 pKa = 4.18PLVGVYY285 pKa = 10.75GDD287 pKa = 3.72DD288 pKa = 3.68VEE290 pKa = 5.05IYY292 pKa = 10.15FYY294 pKa = 11.6ANKK297 pKa = 10.09KK298 pKa = 10.07LYY300 pKa = 10.79AVDD303 pKa = 4.13PDD305 pKa = 4.4FKK307 pKa = 11.52QMDD310 pKa = 3.95GSVDD314 pKa = 3.94LVTVKK319 pKa = 10.57AVCGYY324 pKa = 10.3INYY327 pKa = 9.82AFEE330 pKa = 4.57RR331 pKa = 11.84KK332 pKa = 8.87WGEE335 pKa = 3.64NQLWRR340 pKa = 11.84EE341 pKa = 3.97VLKK344 pKa = 11.0LLSKK348 pKa = 10.73LATSPPMIIHH358 pKa = 6.53GSKK361 pKa = 10.2VFRR364 pKa = 11.84KK365 pKa = 9.28KK366 pKa = 10.67SSGGIISGVVGTTLFDD382 pKa = 3.48TAKK385 pKa = 10.71SIVAYY390 pKa = 10.02HH391 pKa = 6.6HH392 pKa = 6.59LLEE395 pKa = 5.07HH396 pKa = 5.75VQHH399 pKa = 7.55DD400 pKa = 4.3YY401 pKa = 11.06TLLLDD406 pKa = 5.24AGFCKK411 pKa = 10.25QFLKK415 pKa = 10.15QHH417 pKa = 6.4HH418 pKa = 6.26ALEE421 pKa = 4.63VKK423 pKa = 9.95QDD425 pKa = 2.79SWNPVEE431 pKa = 5.43VNMNPVHH438 pKa = 7.01LEE440 pKa = 3.69PFVEE444 pKa = 4.4NKK446 pKa = 9.99FLGVMRR452 pKa = 11.84YY453 pKa = 7.89WYY455 pKa = 9.85EE456 pKa = 4.03LDD458 pKa = 4.1DD459 pKa = 3.73STFIIVPYY467 pKa = 9.99LHH469 pKa = 6.37EE470 pKa = 5.5HH471 pKa = 6.88DD472 pKa = 3.8WLTNILAPRR481 pKa = 11.84TVDD484 pKa = 3.64SISGPTRR491 pKa = 11.84KK492 pKa = 9.61PSDD495 pKa = 3.42MTSEE499 pKa = 4.65RR500 pKa = 11.84ISFDD504 pKa = 3.07RR505 pKa = 11.84ARR507 pKa = 11.84GLLVTGGIFNKK518 pKa = 10.11KK519 pKa = 9.56IKK521 pKa = 10.24DD522 pKa = 3.71ALYY525 pKa = 10.44CAIDD529 pKa = 4.82KK530 pKa = 10.73IPSLPIVMHH539 pKa = 5.43VQAGGGRR546 pKa = 11.84GEE548 pKa = 4.84KK549 pKa = 10.04PGEE552 pKa = 4.14TNVVGNNFSFTDD564 pKa = 3.13SHH566 pKa = 6.77MIPSVGFVEE575 pKa = 6.06DD576 pKa = 3.78IYY578 pKa = 11.33TPIHH582 pKa = 6.26RR583 pKa = 11.84RR584 pKa = 11.84KK585 pKa = 9.53GHH587 pKa = 6.13PPVYY591 pKa = 10.13LYY593 pKa = 7.6PTLVEE598 pKa = 3.94QLNLSRR604 pKa = 11.84NKK606 pKa = 10.03NRR608 pKa = 11.84LVEE611 pKa = 3.99AFKK614 pKa = 10.9CFKK617 pKa = 10.89VVDD620 pKa = 3.91NTTTRR625 pKa = 11.84HH626 pKa = 5.09VKK628 pKa = 8.93GQYY631 pKa = 9.12VKK633 pKa = 10.17PVEE636 pKa = 4.28RR637 pKa = 11.84NLEE640 pKa = 4.35SKK642 pKa = 10.88VPEE645 pKa = 3.9HH646 pKa = 7.28VNFSKK651 pKa = 11.12LNLPFAFGKK660 pKa = 9.39TSGAKK665 pKa = 9.64PLQQLSDD672 pKa = 3.75YY673 pKa = 9.83AYY675 pKa = 8.76HH676 pKa = 6.13HH677 pKa = 5.81TVEE680 pKa = 5.12FGPMPKK686 pKa = 10.02SFVSQFVPIPQNLAPKK702 pKa = 10.5APILQAYY709 pKa = 8.46AKK711 pKa = 10.39LNLVPDD717 pKa = 4.34GWNEE721 pKa = 3.75TNIRR725 pKa = 11.84KK726 pKa = 9.7SKK728 pKa = 10.68LSSLYY733 pKa = 11.04NKK735 pKa = 10.42DD736 pKa = 2.55ITYY739 pKa = 10.1KK740 pKa = 10.58HH741 pKa = 6.08AVEE744 pKa = 5.33KK745 pKa = 10.16IDD747 pKa = 3.97RR748 pKa = 11.84LIKK751 pKa = 10.86ARR753 pKa = 11.84DD754 pKa = 3.53TPPAVDD760 pKa = 3.58VTPQLLIEE768 pKa = 4.45GVVDD772 pKa = 2.9IHH774 pKa = 7.67VEE776 pKa = 3.92NVKK779 pKa = 10.37RR780 pKa = 11.84AFVVYY785 pKa = 9.64PLYY788 pKa = 10.85EE789 pKa = 4.19DD790 pKa = 4.36TGLHH794 pKa = 5.45WLSSIFQQNSLKK806 pKa = 10.55KK807 pKa = 8.68RR808 pKa = 11.84WSTEE812 pKa = 3.4VTEE815 pKa = 4.44SKK817 pKa = 10.42KK818 pKa = 10.38VQLSAEE824 pKa = 4.24SLQGAVIPLARR835 pKa = 11.84VTGKK839 pKa = 10.34DD840 pKa = 3.02LAASLKK846 pKa = 10.8LKK848 pKa = 10.19IAQEE852 pKa = 3.88LFNRR856 pKa = 11.84LRR858 pKa = 11.84AEE860 pKa = 4.38PDD862 pKa = 3.27ITWTPVGKK870 pKa = 9.83KK871 pKa = 9.51GKK873 pKa = 10.28RR874 pKa = 11.84IGEE877 pKa = 4.39SVANTSDD884 pKa = 3.27NVNVAEE890 pKa = 5.03GPIKK894 pKa = 10.51DD895 pKa = 3.78WSNMIEE901 pKa = 4.39TINPSDD907 pKa = 3.64MVVEE911 pKa = 4.27DD912 pKa = 4.6FKK914 pKa = 11.65KK915 pKa = 10.81LEE917 pKa = 4.22YY918 pKa = 10.61YY919 pKa = 11.16SNLLKK924 pKa = 10.21TKK926 pKa = 10.15LKK928 pKa = 10.14QFEE931 pKa = 4.59EE932 pKa = 4.26INKK935 pKa = 9.74KK936 pKa = 10.5DD937 pKa = 3.46EE938 pKa = 4.48NKK940 pKa = 10.74NKK942 pKa = 10.04TEE944 pKa = 3.82FDD946 pKa = 3.05QTRR949 pKa = 11.84RR950 pKa = 11.84NFGGVNEE957 pKa = 5.09SSNQSKK963 pKa = 10.24NRR965 pKa = 11.84LSGARR970 pKa = 11.84KK971 pKa = 8.26KK972 pKa = 8.91TSKK975 pKa = 10.42KK976 pKa = 9.31KK977 pKa = 7.7EE978 pKa = 3.75QRR980 pKa = 11.84EE981 pKa = 4.01VSTGGNANASTEE993 pKa = 4.39FRR995 pKa = 11.84FNWII999 pKa = 3.36
MM1 pKa = 7.32MNMRR5 pKa = 11.84LNPSSEE11 pKa = 4.02PLDD14 pKa = 3.81VQQLDD19 pKa = 3.95FLTEE23 pKa = 4.04LSPFPVFNLQNAQKK37 pKa = 9.01MQIHH41 pKa = 6.37PPKK44 pKa = 10.86VNMTGGFVSVARR56 pKa = 11.84SFGKK60 pKa = 9.87EE61 pKa = 3.29AARR64 pKa = 11.84DD65 pKa = 3.53RR66 pKa = 11.84RR67 pKa = 11.84LLGGALAYY75 pKa = 7.6FTQNEE80 pKa = 4.23VAEE83 pKa = 4.37ILQKK87 pKa = 9.64TVFCYY92 pKa = 9.35GTAKK96 pKa = 10.67GFLTRR101 pKa = 11.84LKK103 pKa = 10.59EE104 pKa = 3.92LTTRR108 pKa = 11.84DD109 pKa = 3.62CKK111 pKa = 10.48TPYY114 pKa = 10.25SISQTVSILNKK125 pKa = 9.95RR126 pKa = 11.84MPVLAEE132 pKa = 4.4EE133 pKa = 4.46LPSWEE138 pKa = 4.9DD139 pKa = 3.38FLSLLEE145 pKa = 4.37NTDD148 pKa = 3.38LTYY151 pKa = 10.67SASAGPPYY159 pKa = 9.71WRR161 pKa = 11.84SKK163 pKa = 10.95LMSLEE168 pKa = 4.09DD169 pKa = 3.68MEE171 pKa = 6.18LEE173 pKa = 4.13VLPLVLEE180 pKa = 4.17HH181 pKa = 6.36MKK183 pKa = 10.82AGTLNQLYY191 pKa = 9.64HH192 pKa = 6.29LHH194 pKa = 7.14PEE196 pKa = 4.29LFLVEE201 pKa = 4.67IKK203 pKa = 10.98NKK205 pKa = 9.72LDD207 pKa = 3.42RR208 pKa = 11.84YY209 pKa = 7.93EE210 pKa = 3.95TAKK213 pKa = 10.61FDD215 pKa = 4.16VKK217 pKa = 10.47CRR219 pKa = 11.84PYY221 pKa = 11.27GNVPFHH227 pKa = 6.41MGFLASMIVQPFCKK241 pKa = 10.01KK242 pKa = 10.4LKK244 pKa = 9.58LWHH247 pKa = 6.9NSDD250 pKa = 2.71SWNAYY255 pKa = 8.48GFSFAEE261 pKa = 4.41GSYY264 pKa = 10.95LEE266 pKa = 4.52LYY268 pKa = 9.91EE269 pKa = 4.2RR270 pKa = 11.84LKK272 pKa = 10.7KK273 pKa = 10.58FKK275 pKa = 10.71RR276 pKa = 11.84GDD278 pKa = 3.45EE279 pKa = 4.18PLVGVYY285 pKa = 10.75GDD287 pKa = 3.72DD288 pKa = 3.68VEE290 pKa = 5.05IYY292 pKa = 10.15FYY294 pKa = 11.6ANKK297 pKa = 10.09KK298 pKa = 10.07LYY300 pKa = 10.79AVDD303 pKa = 4.13PDD305 pKa = 4.4FKK307 pKa = 11.52QMDD310 pKa = 3.95GSVDD314 pKa = 3.94LVTVKK319 pKa = 10.57AVCGYY324 pKa = 10.3INYY327 pKa = 9.82AFEE330 pKa = 4.57RR331 pKa = 11.84KK332 pKa = 8.87WGEE335 pKa = 3.64NQLWRR340 pKa = 11.84EE341 pKa = 3.97VLKK344 pKa = 11.0LLSKK348 pKa = 10.73LATSPPMIIHH358 pKa = 6.53GSKK361 pKa = 10.2VFRR364 pKa = 11.84KK365 pKa = 9.28KK366 pKa = 10.67SSGGIISGVVGTTLFDD382 pKa = 3.48TAKK385 pKa = 10.71SIVAYY390 pKa = 10.02HH391 pKa = 6.6HH392 pKa = 6.59LLEE395 pKa = 5.07HH396 pKa = 5.75VQHH399 pKa = 7.55DD400 pKa = 4.3YY401 pKa = 11.06TLLLDD406 pKa = 5.24AGFCKK411 pKa = 10.25QFLKK415 pKa = 10.15QHH417 pKa = 6.4HH418 pKa = 6.26ALEE421 pKa = 4.63VKK423 pKa = 9.95QDD425 pKa = 2.79SWNPVEE431 pKa = 5.43VNMNPVHH438 pKa = 7.01LEE440 pKa = 3.69PFVEE444 pKa = 4.4NKK446 pKa = 9.99FLGVMRR452 pKa = 11.84YY453 pKa = 7.89WYY455 pKa = 9.85EE456 pKa = 4.03LDD458 pKa = 4.1DD459 pKa = 3.73STFIIVPYY467 pKa = 9.99LHH469 pKa = 6.37EE470 pKa = 5.5HH471 pKa = 6.88DD472 pKa = 3.8WLTNILAPRR481 pKa = 11.84TVDD484 pKa = 3.64SISGPTRR491 pKa = 11.84KK492 pKa = 9.61PSDD495 pKa = 3.42MTSEE499 pKa = 4.65RR500 pKa = 11.84ISFDD504 pKa = 3.07RR505 pKa = 11.84ARR507 pKa = 11.84GLLVTGGIFNKK518 pKa = 10.11KK519 pKa = 9.56IKK521 pKa = 10.24DD522 pKa = 3.71ALYY525 pKa = 10.44CAIDD529 pKa = 4.82KK530 pKa = 10.73IPSLPIVMHH539 pKa = 5.43VQAGGGRR546 pKa = 11.84GEE548 pKa = 4.84KK549 pKa = 10.04PGEE552 pKa = 4.14TNVVGNNFSFTDD564 pKa = 3.13SHH566 pKa = 6.77MIPSVGFVEE575 pKa = 6.06DD576 pKa = 3.78IYY578 pKa = 11.33TPIHH582 pKa = 6.26RR583 pKa = 11.84RR584 pKa = 11.84KK585 pKa = 9.53GHH587 pKa = 6.13PPVYY591 pKa = 10.13LYY593 pKa = 7.6PTLVEE598 pKa = 3.94QLNLSRR604 pKa = 11.84NKK606 pKa = 10.03NRR608 pKa = 11.84LVEE611 pKa = 3.99AFKK614 pKa = 10.9CFKK617 pKa = 10.89VVDD620 pKa = 3.91NTTTRR625 pKa = 11.84HH626 pKa = 5.09VKK628 pKa = 8.93GQYY631 pKa = 9.12VKK633 pKa = 10.17PVEE636 pKa = 4.28RR637 pKa = 11.84NLEE640 pKa = 4.35SKK642 pKa = 10.88VPEE645 pKa = 3.9HH646 pKa = 7.28VNFSKK651 pKa = 11.12LNLPFAFGKK660 pKa = 9.39TSGAKK665 pKa = 9.64PLQQLSDD672 pKa = 3.75YY673 pKa = 9.83AYY675 pKa = 8.76HH676 pKa = 6.13HH677 pKa = 5.81TVEE680 pKa = 5.12FGPMPKK686 pKa = 10.02SFVSQFVPIPQNLAPKK702 pKa = 10.5APILQAYY709 pKa = 8.46AKK711 pKa = 10.39LNLVPDD717 pKa = 4.34GWNEE721 pKa = 3.75TNIRR725 pKa = 11.84KK726 pKa = 9.7SKK728 pKa = 10.68LSSLYY733 pKa = 11.04NKK735 pKa = 10.42DD736 pKa = 2.55ITYY739 pKa = 10.1KK740 pKa = 10.58HH741 pKa = 6.08AVEE744 pKa = 5.33KK745 pKa = 10.16IDD747 pKa = 3.97RR748 pKa = 11.84LIKK751 pKa = 10.86ARR753 pKa = 11.84DD754 pKa = 3.53TPPAVDD760 pKa = 3.58VTPQLLIEE768 pKa = 4.45GVVDD772 pKa = 2.9IHH774 pKa = 7.67VEE776 pKa = 3.92NVKK779 pKa = 10.37RR780 pKa = 11.84AFVVYY785 pKa = 9.64PLYY788 pKa = 10.85EE789 pKa = 4.19DD790 pKa = 4.36TGLHH794 pKa = 5.45WLSSIFQQNSLKK806 pKa = 10.55KK807 pKa = 8.68RR808 pKa = 11.84WSTEE812 pKa = 3.4VTEE815 pKa = 4.44SKK817 pKa = 10.42KK818 pKa = 10.38VQLSAEE824 pKa = 4.24SLQGAVIPLARR835 pKa = 11.84VTGKK839 pKa = 10.34DD840 pKa = 3.02LAASLKK846 pKa = 10.8LKK848 pKa = 10.19IAQEE852 pKa = 3.88LFNRR856 pKa = 11.84LRR858 pKa = 11.84AEE860 pKa = 4.38PDD862 pKa = 3.27ITWTPVGKK870 pKa = 9.83KK871 pKa = 9.51GKK873 pKa = 10.28RR874 pKa = 11.84IGEE877 pKa = 4.39SVANTSDD884 pKa = 3.27NVNVAEE890 pKa = 5.03GPIKK894 pKa = 10.51DD895 pKa = 3.78WSNMIEE901 pKa = 4.39TINPSDD907 pKa = 3.64MVVEE911 pKa = 4.27DD912 pKa = 4.6FKK914 pKa = 11.65KK915 pKa = 10.81LEE917 pKa = 4.22YY918 pKa = 10.61YY919 pKa = 11.16SNLLKK924 pKa = 10.21TKK926 pKa = 10.15LKK928 pKa = 10.14QFEE931 pKa = 4.59EE932 pKa = 4.26INKK935 pKa = 9.74KK936 pKa = 10.5DD937 pKa = 3.46EE938 pKa = 4.48NKK940 pKa = 10.74NKK942 pKa = 10.04TEE944 pKa = 3.82FDD946 pKa = 3.05QTRR949 pKa = 11.84RR950 pKa = 11.84NFGGVNEE957 pKa = 5.09SSNQSKK963 pKa = 10.24NRR965 pKa = 11.84LSGARR970 pKa = 11.84KK971 pKa = 8.26KK972 pKa = 8.91TSKK975 pKa = 10.42KK976 pKa = 9.31KK977 pKa = 7.7EE978 pKa = 3.75QRR980 pKa = 11.84EE981 pKa = 4.01VSTGGNANASTEE993 pKa = 4.39FRR995 pKa = 11.84FNWII999 pKa = 3.36
Molecular weight: 113.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2RVN9|A0A1B2RVN9_9VIRU RNA-dependent RNA polymerase OS=Egaro virus OX=1888318 PE=4 SV=1
MM1 pKa = 6.92KK2 pKa = 9.99TKK4 pKa = 10.06TKK6 pKa = 10.68QNLIKK11 pKa = 10.22QDD13 pKa = 3.46GTLEE17 pKa = 4.11EE18 pKa = 4.35LMKK21 pKa = 10.93AQIKK25 pKa = 10.43AKK27 pKa = 9.5IASLAPVKK35 pKa = 10.23RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.39RR40 pKa = 11.84RR41 pKa = 11.84NKK43 pKa = 10.34GKK45 pKa = 9.85FPPVAMPMPQRR56 pKa = 11.84SFALTGFNKK65 pKa = 10.55DD66 pKa = 3.23EE67 pKa = 4.26GVDD70 pKa = 3.97RR71 pKa = 11.84IFHH74 pKa = 6.45IPDD77 pKa = 2.93ISKK80 pKa = 10.79DD81 pKa = 3.46IVDD84 pKa = 3.88KK85 pKa = 11.19QILIDD90 pKa = 3.97IPVLPTLGSRR100 pKa = 11.84LGIIASAWQRR110 pKa = 11.84IVYY113 pKa = 9.75KK114 pKa = 10.35KK115 pKa = 10.8LEE117 pKa = 3.98FSITPKK123 pKa = 10.23TSTAVSGGYY132 pKa = 9.01IAAFIPDD139 pKa = 3.73VTDD142 pKa = 3.27VLTGTSPDD150 pKa = 3.41FALDD154 pKa = 3.64KK155 pKa = 11.07LSSQPSAVEE164 pKa = 3.98AKK166 pKa = 7.96WWEE169 pKa = 4.14SSTVRR174 pKa = 11.84VPSLPDD180 pKa = 3.32LFYY183 pKa = 10.52TSPSPEE189 pKa = 3.98SPRR192 pKa = 11.84FSSPGRR198 pKa = 11.84FVLIVDD204 pKa = 4.71GAASQAGSLTIKK216 pKa = 8.08MHH218 pKa = 5.77WSVLLSEE225 pKa = 5.17PSLEE229 pKa = 4.37GNNAPADD236 pKa = 3.52QKK238 pKa = 11.46EE239 pKa = 4.08FVIKK243 pKa = 10.7DD244 pKa = 4.02GLWMQAKK251 pKa = 10.32AGGIFVNTSSDD262 pKa = 3.34NTKK265 pKa = 10.54KK266 pKa = 10.74VLNSNVLLVLPDD278 pKa = 3.41AKK280 pKa = 10.62EE281 pKa = 3.8NQIYY285 pKa = 10.65RR286 pKa = 11.84MATTASYY293 pKa = 10.37LRR295 pKa = 11.84CADD298 pKa = 3.63TVTLDD303 pKa = 4.03NIWLRR308 pKa = 11.84SFWYY312 pKa = 9.39FIIVKK317 pKa = 8.8TSSGGLQMYY326 pKa = 8.69PYY328 pKa = 9.57TPEE331 pKa = 3.7NKK333 pKa = 9.95EE334 pKa = 3.78ILKK337 pKa = 10.79DD338 pKa = 3.61NSAGDD343 pKa = 4.07TEE345 pKa = 4.61VLPAGHH351 pKa = 6.48EE352 pKa = 4.2VLLEE356 pKa = 3.86STAQVDD362 pKa = 4.48FPRR365 pKa = 11.84GSEE368 pKa = 4.31FICASDD374 pKa = 3.47RR375 pKa = 11.84KK376 pKa = 10.38SLLHH380 pKa = 6.57HH381 pKa = 7.11FYY383 pKa = 11.25QMSLSLNKK391 pKa = 9.65NRR393 pKa = 11.84QQYY396 pKa = 7.31QTASSNKK403 pKa = 9.34CLTFLPLYY411 pKa = 10.55SKK413 pKa = 10.57FISSLKK419 pKa = 10.37RR420 pKa = 11.84EE421 pKa = 4.08DD422 pKa = 3.52SFGEE426 pKa = 4.05EE427 pKa = 3.57KK428 pKa = 10.66ALRR431 pKa = 11.84WRR433 pKa = 11.84AGSQASFLDD442 pKa = 3.95WEE444 pKa = 4.49TLPEE448 pKa = 4.15KK449 pKa = 9.98LTRR452 pKa = 11.84LEE454 pKa = 4.14VSDD457 pKa = 4.0VQSLKK462 pKa = 11.09
MM1 pKa = 6.92KK2 pKa = 9.99TKK4 pKa = 10.06TKK6 pKa = 10.68QNLIKK11 pKa = 10.22QDD13 pKa = 3.46GTLEE17 pKa = 4.11EE18 pKa = 4.35LMKK21 pKa = 10.93AQIKK25 pKa = 10.43AKK27 pKa = 9.5IASLAPVKK35 pKa = 10.23RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.39RR40 pKa = 11.84RR41 pKa = 11.84NKK43 pKa = 10.34GKK45 pKa = 9.85FPPVAMPMPQRR56 pKa = 11.84SFALTGFNKK65 pKa = 10.55DD66 pKa = 3.23EE67 pKa = 4.26GVDD70 pKa = 3.97RR71 pKa = 11.84IFHH74 pKa = 6.45IPDD77 pKa = 2.93ISKK80 pKa = 10.79DD81 pKa = 3.46IVDD84 pKa = 3.88KK85 pKa = 11.19QILIDD90 pKa = 3.97IPVLPTLGSRR100 pKa = 11.84LGIIASAWQRR110 pKa = 11.84IVYY113 pKa = 9.75KK114 pKa = 10.35KK115 pKa = 10.8LEE117 pKa = 3.98FSITPKK123 pKa = 10.23TSTAVSGGYY132 pKa = 9.01IAAFIPDD139 pKa = 3.73VTDD142 pKa = 3.27VLTGTSPDD150 pKa = 3.41FALDD154 pKa = 3.64KK155 pKa = 11.07LSSQPSAVEE164 pKa = 3.98AKK166 pKa = 7.96WWEE169 pKa = 4.14SSTVRR174 pKa = 11.84VPSLPDD180 pKa = 3.32LFYY183 pKa = 10.52TSPSPEE189 pKa = 3.98SPRR192 pKa = 11.84FSSPGRR198 pKa = 11.84FVLIVDD204 pKa = 4.71GAASQAGSLTIKK216 pKa = 8.08MHH218 pKa = 5.77WSVLLSEE225 pKa = 5.17PSLEE229 pKa = 4.37GNNAPADD236 pKa = 3.52QKK238 pKa = 11.46EE239 pKa = 4.08FVIKK243 pKa = 10.7DD244 pKa = 4.02GLWMQAKK251 pKa = 10.32AGGIFVNTSSDD262 pKa = 3.34NTKK265 pKa = 10.54KK266 pKa = 10.74VLNSNVLLVLPDD278 pKa = 3.41AKK280 pKa = 10.62EE281 pKa = 3.8NQIYY285 pKa = 10.65RR286 pKa = 11.84MATTASYY293 pKa = 10.37LRR295 pKa = 11.84CADD298 pKa = 3.63TVTLDD303 pKa = 4.03NIWLRR308 pKa = 11.84SFWYY312 pKa = 9.39FIIVKK317 pKa = 8.8TSSGGLQMYY326 pKa = 8.69PYY328 pKa = 9.57TPEE331 pKa = 3.7NKK333 pKa = 9.95EE334 pKa = 3.78ILKK337 pKa = 10.79DD338 pKa = 3.61NSAGDD343 pKa = 4.07TEE345 pKa = 4.61VLPAGHH351 pKa = 6.48EE352 pKa = 4.2VLLEE356 pKa = 3.86STAQVDD362 pKa = 4.48FPRR365 pKa = 11.84GSEE368 pKa = 4.31FICASDD374 pKa = 3.47RR375 pKa = 11.84KK376 pKa = 10.38SLLHH380 pKa = 6.57HH381 pKa = 7.11FYY383 pKa = 11.25QMSLSLNKK391 pKa = 9.65NRR393 pKa = 11.84QQYY396 pKa = 7.31QTASSNKK403 pKa = 9.34CLTFLPLYY411 pKa = 10.55SKK413 pKa = 10.57FISSLKK419 pKa = 10.37RR420 pKa = 11.84EE421 pKa = 4.08DD422 pKa = 3.52SFGEE426 pKa = 4.05EE427 pKa = 3.57KK428 pKa = 10.66ALRR431 pKa = 11.84WRR433 pKa = 11.84AGSQASFLDD442 pKa = 3.95WEE444 pKa = 4.49TLPEE448 pKa = 4.15KK449 pKa = 9.98LTRR452 pKa = 11.84LEE454 pKa = 4.14VSDD457 pKa = 4.0VQSLKK462 pKa = 11.09
Molecular weight: 51.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1461 |
462 |
999 |
730.5 |
82.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.955 ± 0.658 | 0.753 ± 0.057 |
5.065 ± 0.312 | 6.023 ± 0.579 |
4.791 ± 0.136 | 5.407 ± 0.238 |
2.19 ± 0.614 | 4.86 ± 0.545 |
8.487 ± 0.265 | 10.062 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.053 ± 0.058 | 4.928 ± 0.811 |
5.613 ± 0.128 | 3.628 ± 0.269 |
4.517 ± 0.135 | 8.008 ± 1.439 |
5.544 ± 0.286 | 7.255 ± 0.781 |
1.643 ± 0.169 | 3.217 ± 0.463 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
