
Halorhodospira halophila (strain DSM 244 / SL1) (Ectothiorhodospira halophila (strain DSM 244 / SL1))
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Halorhodospira; Halorhodospira halophila
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
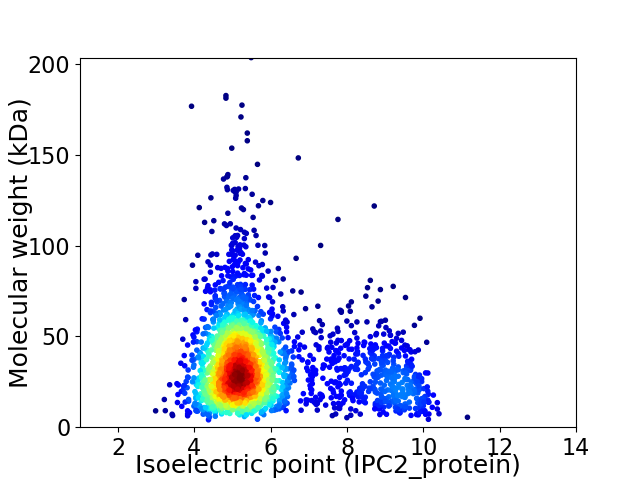
Virtual 2D-PAGE plot for 2406 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A1WWW0|HFLD_HALHL High frequency lysogenization protein HflD homolog OS=Halorhodospira halophila (strain DSM 244 / SL1) OX=349124 GN=hflD PE=3 SV=1
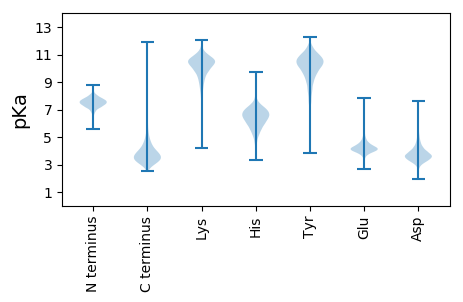
MM1 pKa = 7.87RR2 pKa = 11.84LTRR5 pKa = 11.84RR6 pKa = 11.84AVYY9 pKa = 8.35FAPCLLALGGIATSAHH25 pKa = 5.59GAAFQLQEE33 pKa = 3.95QSASLLASAFAGRR46 pKa = 11.84SADD49 pKa = 3.63AGDD52 pKa = 3.57ISHH55 pKa = 6.68MFFNPATLGVHH66 pKa = 7.16AGRR69 pKa = 11.84GPEE72 pKa = 3.91AEE74 pKa = 4.65LVLSYY79 pKa = 10.65IDD81 pKa = 3.39PTFDD85 pKa = 4.31FEE87 pKa = 5.99ADD89 pKa = 3.56SDD91 pKa = 4.44PGWSAVGGDD100 pKa = 3.83TGQLSASGGEE110 pKa = 4.19SALAPVFYY118 pKa = 10.51AGFDD122 pKa = 3.66LAPDD126 pKa = 3.98LRR128 pKa = 11.84AGLGINVPYY137 pKa = 10.51GLEE140 pKa = 3.75TDD142 pKa = 4.2YY143 pKa = 11.72PEE145 pKa = 4.79DD146 pKa = 3.08WVGRR150 pKa = 11.84YY151 pKa = 9.23DD152 pKa = 4.75AVNTDD157 pKa = 4.09LVTVDD162 pKa = 4.75INPQLGWRR170 pKa = 11.84LDD172 pKa = 3.57EE173 pKa = 4.21QLALGLGVSAQYY185 pKa = 11.74ADD187 pKa = 3.68ATLSTMAPEE196 pKa = 4.68TDD198 pKa = 4.24FSTTPATPTPTPTPANDD215 pKa = 3.52GKK217 pKa = 9.87LTVDD221 pKa = 3.67GDD223 pKa = 3.58SWSYY227 pKa = 11.34SFNLGAHH234 pKa = 5.5YY235 pKa = 10.57AVTEE239 pKa = 4.03DD240 pKa = 3.57TQLGVAYY247 pKa = 10.25RR248 pKa = 11.84HH249 pKa = 5.9GLGHH253 pKa = 6.0TLSGEE258 pKa = 4.12AEE260 pKa = 4.16LVDD263 pKa = 4.43DD264 pKa = 5.07GGVTVDD270 pKa = 3.68EE271 pKa = 4.8TGGEE275 pKa = 4.01ADD277 pKa = 4.2LDD279 pKa = 3.86IPASLNLGVSHH290 pKa = 6.72QLDD293 pKa = 4.08PQWTLLADD301 pKa = 3.86ATWTQWSDD309 pKa = 3.55FEE311 pKa = 4.2EE312 pKa = 4.28LAIEE316 pKa = 4.22FDD318 pKa = 4.12DD319 pKa = 5.72GIVGIPQGQNSYY331 pKa = 11.38AVTDD335 pKa = 3.63EE336 pKa = 4.17QAYY339 pKa = 10.1EE340 pKa = 4.05EE341 pKa = 4.7YY342 pKa = 10.61DD343 pKa = 3.39WDD345 pKa = 4.14DD346 pKa = 2.94TWFLSLGTRR355 pKa = 11.84YY356 pKa = 9.88DD357 pKa = 4.6ASEE360 pKa = 3.96QWDD363 pKa = 3.9LRR365 pKa = 11.84AGLAYY370 pKa = 10.41DD371 pKa = 3.46QTPTGDD377 pKa = 3.31SNRR380 pKa = 11.84TPRR383 pKa = 11.84IPDD386 pKa = 3.32EE387 pKa = 4.42DD388 pKa = 4.12RR389 pKa = 11.84TWLAVGGTWSPAQADD404 pKa = 3.65QLRR407 pKa = 11.84VDD409 pKa = 4.49FGYY412 pKa = 9.54TYY414 pKa = 10.67IWLDD418 pKa = 3.53DD419 pKa = 3.82ADD421 pKa = 4.13IEE423 pKa = 4.43VAEE426 pKa = 4.83DD427 pKa = 3.61FGGVTGEE434 pKa = 4.28YY435 pKa = 9.68EE436 pKa = 4.41SNVQVFTTSANWRR449 pKa = 11.84FF450 pKa = 3.38
MM1 pKa = 7.87RR2 pKa = 11.84LTRR5 pKa = 11.84RR6 pKa = 11.84AVYY9 pKa = 8.35FAPCLLALGGIATSAHH25 pKa = 5.59GAAFQLQEE33 pKa = 3.95QSASLLASAFAGRR46 pKa = 11.84SADD49 pKa = 3.63AGDD52 pKa = 3.57ISHH55 pKa = 6.68MFFNPATLGVHH66 pKa = 7.16AGRR69 pKa = 11.84GPEE72 pKa = 3.91AEE74 pKa = 4.65LVLSYY79 pKa = 10.65IDD81 pKa = 3.39PTFDD85 pKa = 4.31FEE87 pKa = 5.99ADD89 pKa = 3.56SDD91 pKa = 4.44PGWSAVGGDD100 pKa = 3.83TGQLSASGGEE110 pKa = 4.19SALAPVFYY118 pKa = 10.51AGFDD122 pKa = 3.66LAPDD126 pKa = 3.98LRR128 pKa = 11.84AGLGINVPYY137 pKa = 10.51GLEE140 pKa = 3.75TDD142 pKa = 4.2YY143 pKa = 11.72PEE145 pKa = 4.79DD146 pKa = 3.08WVGRR150 pKa = 11.84YY151 pKa = 9.23DD152 pKa = 4.75AVNTDD157 pKa = 4.09LVTVDD162 pKa = 4.75INPQLGWRR170 pKa = 11.84LDD172 pKa = 3.57EE173 pKa = 4.21QLALGLGVSAQYY185 pKa = 11.74ADD187 pKa = 3.68ATLSTMAPEE196 pKa = 4.68TDD198 pKa = 4.24FSTTPATPTPTPTPANDD215 pKa = 3.52GKK217 pKa = 9.87LTVDD221 pKa = 3.67GDD223 pKa = 3.58SWSYY227 pKa = 11.34SFNLGAHH234 pKa = 5.5YY235 pKa = 10.57AVTEE239 pKa = 4.03DD240 pKa = 3.57TQLGVAYY247 pKa = 10.25RR248 pKa = 11.84HH249 pKa = 5.9GLGHH253 pKa = 6.0TLSGEE258 pKa = 4.12AEE260 pKa = 4.16LVDD263 pKa = 4.43DD264 pKa = 5.07GGVTVDD270 pKa = 3.68EE271 pKa = 4.8TGGEE275 pKa = 4.01ADD277 pKa = 4.2LDD279 pKa = 3.86IPASLNLGVSHH290 pKa = 6.72QLDD293 pKa = 4.08PQWTLLADD301 pKa = 3.86ATWTQWSDD309 pKa = 3.55FEE311 pKa = 4.2EE312 pKa = 4.28LAIEE316 pKa = 4.22FDD318 pKa = 4.12DD319 pKa = 5.72GIVGIPQGQNSYY331 pKa = 11.38AVTDD335 pKa = 3.63EE336 pKa = 4.17QAYY339 pKa = 10.1EE340 pKa = 4.05EE341 pKa = 4.7YY342 pKa = 10.61DD343 pKa = 3.39WDD345 pKa = 4.14DD346 pKa = 2.94TWFLSLGTRR355 pKa = 11.84YY356 pKa = 9.88DD357 pKa = 4.6ASEE360 pKa = 3.96QWDD363 pKa = 3.9LRR365 pKa = 11.84AGLAYY370 pKa = 10.41DD371 pKa = 3.46QTPTGDD377 pKa = 3.31SNRR380 pKa = 11.84TPRR383 pKa = 11.84IPDD386 pKa = 3.32EE387 pKa = 4.42DD388 pKa = 4.12RR389 pKa = 11.84TWLAVGGTWSPAQADD404 pKa = 3.65QLRR407 pKa = 11.84VDD409 pKa = 4.49FGYY412 pKa = 9.54TYY414 pKa = 10.67IWLDD418 pKa = 3.53DD419 pKa = 3.82ADD421 pKa = 4.13IEE423 pKa = 4.43VAEE426 pKa = 4.83DD427 pKa = 3.61FGGVTGEE434 pKa = 4.28YY435 pKa = 9.68EE436 pKa = 4.41SNVQVFTTSANWRR449 pKa = 11.84FF450 pKa = 3.38
Molecular weight: 48.47 kDa
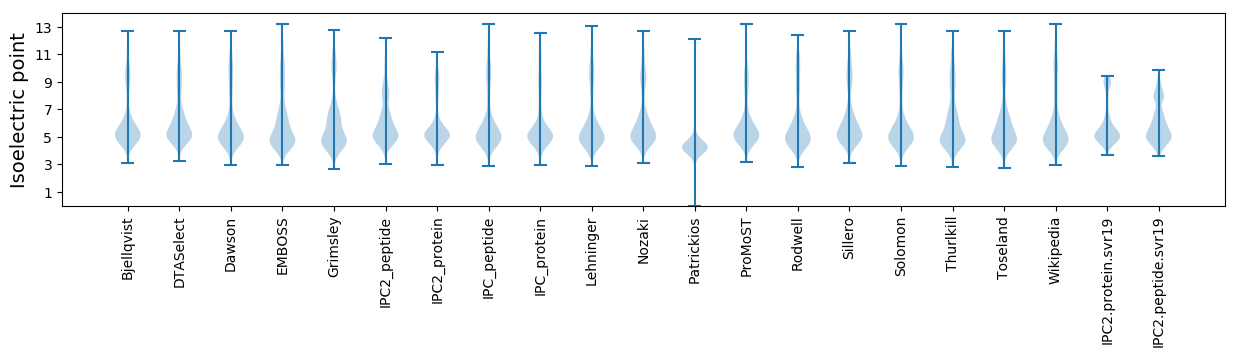
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A1WWG0|A1WWG0_HALHL Biopolymer transport protein ExbD/TolR OS=Halorhodospira halophila (strain DSM 244 / SL1) OX=349124 GN=Hhal_1247 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.19QPSNIKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 7.88RR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84LVLKK32 pKa = 10.32RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 9.0RR41 pKa = 11.84LTPTAA46 pKa = 4.02
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.19QPSNIKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 7.88RR14 pKa = 11.84THH16 pKa = 5.91GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84LVLKK32 pKa = 10.32RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 9.0RR41 pKa = 11.84LTPTAA46 pKa = 4.02
Molecular weight: 5.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
815824 |
36 |
1896 |
339.1 |
37.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.968 ± 0.072 | 0.905 ± 0.017 |
6.066 ± 0.039 | 7.489 ± 0.051 |
3.057 ± 0.029 | 8.891 ± 0.046 |
2.45 ± 0.023 | 4.094 ± 0.035 |
1.76 ± 0.038 | 10.72 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.983 ± 0.022 | 2.01 ± 0.024 |
5.381 ± 0.041 | 3.859 ± 0.029 |
8.709 ± 0.056 | 4.521 ± 0.03 |
4.81 ± 0.03 | 7.518 ± 0.041 |
1.434 ± 0.022 | 2.378 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
