
Crucibulum laeve
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Agaricales incertae sedis; Crucibulum
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 14180 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
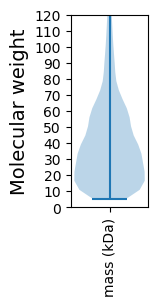
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A5C3LQA1|A0A5C3LQA1_9AGAR Uncharacterized protein OS=Crucibulum laeve OX=68775 GN=BDQ12DRAFT_688331 PE=4 SV=1
MM1 pKa = 7.72SNRR4 pKa = 11.84DD5 pKa = 3.67DD6 pKa = 3.63NSYY9 pKa = 11.53GSGNDD14 pKa = 3.33SYY16 pKa = 12.15GSGNNDD22 pKa = 2.84SYY24 pKa = 11.85GSSNRR29 pKa = 11.84SGDD32 pKa = 3.24NDD34 pKa = 3.42NNNRR38 pKa = 11.84RR39 pKa = 11.84SGDD42 pKa = 3.39DD43 pKa = 3.51DD44 pKa = 3.96NSYY47 pKa = 11.39GSNNNSSTNDD57 pKa = 3.14SYY59 pKa = 12.1GSGRR63 pKa = 11.84DD64 pKa = 3.38RR65 pKa = 11.84SDD67 pKa = 5.16DD68 pKa = 3.61NDD70 pKa = 3.49NTSSGNTYY78 pKa = 10.43GSSNNDD84 pKa = 2.84SYY86 pKa = 11.99GSSNTGSNKK95 pKa = 9.74EE96 pKa = 4.08SGSYY100 pKa = 10.51GSNDD104 pKa = 2.93NSYY107 pKa = 11.15GSSAKK112 pKa = 10.33DD113 pKa = 3.16SYY115 pKa = 11.66GSSNNDD121 pKa = 2.91TYY123 pKa = 11.65GSSNKK128 pKa = 9.24EE129 pKa = 3.62SSYY132 pKa = 11.1GSSSNEE138 pKa = 3.56DD139 pKa = 3.42SYY141 pKa = 12.21GSSKK145 pKa = 10.33KK146 pKa = 10.35DD147 pKa = 3.13SYY149 pKa = 11.79GSNNDD154 pKa = 3.3SYY156 pKa = 11.89GSSNNDD162 pKa = 3.11SNKK165 pKa = 9.33GDD167 pKa = 3.83SYY169 pKa = 11.77GSSNNNRR176 pKa = 11.84SSNEE180 pKa = 3.55DD181 pKa = 3.27SYY183 pKa = 12.16GSNNEE188 pKa = 3.64SSYY191 pKa = 11.54GNNDD195 pKa = 3.26NEE197 pKa = 4.72SSNNNSNNNNNSQGKK212 pKa = 9.61SGDD215 pKa = 3.5WVDD218 pKa = 4.15KK219 pKa = 10.29GVSYY223 pKa = 10.58AAKK226 pKa = 8.81QAGYY230 pKa = 10.46NVDD233 pKa = 3.36EE234 pKa = 4.51ATADD238 pKa = 3.7KK239 pKa = 10.37IGNSISQGLGKK250 pKa = 10.18FVGGGPGGILL260 pKa = 3.44
MM1 pKa = 7.72SNRR4 pKa = 11.84DD5 pKa = 3.67DD6 pKa = 3.63NSYY9 pKa = 11.53GSGNDD14 pKa = 3.33SYY16 pKa = 12.15GSGNNDD22 pKa = 2.84SYY24 pKa = 11.85GSSNRR29 pKa = 11.84SGDD32 pKa = 3.24NDD34 pKa = 3.42NNNRR38 pKa = 11.84RR39 pKa = 11.84SGDD42 pKa = 3.39DD43 pKa = 3.51DD44 pKa = 3.96NSYY47 pKa = 11.39GSNNNSSTNDD57 pKa = 3.14SYY59 pKa = 12.1GSGRR63 pKa = 11.84DD64 pKa = 3.38RR65 pKa = 11.84SDD67 pKa = 5.16DD68 pKa = 3.61NDD70 pKa = 3.49NTSSGNTYY78 pKa = 10.43GSSNNDD84 pKa = 2.84SYY86 pKa = 11.99GSSNTGSNKK95 pKa = 9.74EE96 pKa = 4.08SGSYY100 pKa = 10.51GSNDD104 pKa = 2.93NSYY107 pKa = 11.15GSSAKK112 pKa = 10.33DD113 pKa = 3.16SYY115 pKa = 11.66GSSNNDD121 pKa = 2.91TYY123 pKa = 11.65GSSNKK128 pKa = 9.24EE129 pKa = 3.62SSYY132 pKa = 11.1GSSSNEE138 pKa = 3.56DD139 pKa = 3.42SYY141 pKa = 12.21GSSKK145 pKa = 10.33KK146 pKa = 10.35DD147 pKa = 3.13SYY149 pKa = 11.79GSNNDD154 pKa = 3.3SYY156 pKa = 11.89GSSNNDD162 pKa = 3.11SNKK165 pKa = 9.33GDD167 pKa = 3.83SYY169 pKa = 11.77GSSNNNRR176 pKa = 11.84SSNEE180 pKa = 3.55DD181 pKa = 3.27SYY183 pKa = 12.16GSNNEE188 pKa = 3.64SSYY191 pKa = 11.54GNNDD195 pKa = 3.26NEE197 pKa = 4.72SSNNNSNNNNNSQGKK212 pKa = 9.61SGDD215 pKa = 3.5WVDD218 pKa = 4.15KK219 pKa = 10.29GVSYY223 pKa = 10.58AAKK226 pKa = 8.81QAGYY230 pKa = 10.46NVDD233 pKa = 3.36EE234 pKa = 4.51ATADD238 pKa = 3.7KK239 pKa = 10.37IGNSISQGLGKK250 pKa = 10.18FVGGGPGGILL260 pKa = 3.44
Molecular weight: 26.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C3M7Z2|A0A5C3M7Z2_9AGAR Transmembrane amino acid transporter protein-domain-containing protein OS=Crucibulum laeve OX=68775 GN=BDQ12DRAFT_710842 PE=3 SV=1
MM1 pKa = 7.95LMRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.6KK7 pKa = 10.26RR8 pKa = 11.84NLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.5LPQGRR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.61QKK22 pKa = 10.23LRR24 pKa = 11.84TRR26 pKa = 11.84RR27 pKa = 11.84AARR30 pKa = 11.84RR31 pKa = 11.84VRR33 pKa = 11.84SPRR36 pKa = 11.84QSVPLPKK43 pKa = 9.9KK44 pKa = 9.8RR45 pKa = 11.84LSPRR49 pKa = 11.84KK50 pKa = 8.85RR51 pKa = 11.84LLPRR55 pKa = 11.84SVLPRR60 pKa = 11.84KK61 pKa = 9.44RR62 pKa = 11.84RR63 pKa = 11.84PNQRR67 pKa = 11.84TSLEE71 pKa = 4.23KK72 pKa = 10.26ISPKK76 pKa = 10.23RR77 pKa = 11.84STPCPPIPTTKK88 pKa = 10.55LSLRR92 pKa = 11.84KK93 pKa = 9.43RR94 pKa = 11.84NPRR97 pKa = 11.84KK98 pKa = 9.74RR99 pKa = 11.84PLLRR103 pKa = 11.84RR104 pKa = 11.84SPPRR108 pKa = 11.84KK109 pKa = 9.38LLHH112 pKa = 5.7LRR114 pKa = 11.84LRR116 pKa = 11.84KK117 pKa = 9.29RR118 pKa = 11.84RR119 pKa = 11.84LSLRR123 pKa = 11.84RR124 pKa = 11.84KK125 pKa = 7.9QPQRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84MRR133 pKa = 11.84KK134 pKa = 9.18RR135 pKa = 11.84MSIWRR140 pKa = 11.84MPLSTKK146 pKa = 9.25RR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84VVASARR155 pKa = 11.84SLHH158 pKa = 6.75RR159 pKa = 11.84KK160 pKa = 8.59RR161 pKa = 11.84QLSLPRR167 pKa = 11.84RR168 pKa = 11.84KK169 pKa = 9.64PNPPPRR175 pKa = 11.84QRR177 pKa = 11.84RR178 pKa = 11.84ARR180 pKa = 11.84KK181 pKa = 8.7LLRR184 pKa = 11.84MMEE187 pKa = 4.3TMLMTDD193 pKa = 3.96DD194 pKa = 4.3YY195 pKa = 11.31LPVVPLQCFISFDD208 pKa = 3.42
MM1 pKa = 7.95LMRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.6KK7 pKa = 10.26RR8 pKa = 11.84NLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 9.5LPQGRR18 pKa = 11.84RR19 pKa = 11.84KK20 pKa = 9.61QKK22 pKa = 10.23LRR24 pKa = 11.84TRR26 pKa = 11.84RR27 pKa = 11.84AARR30 pKa = 11.84RR31 pKa = 11.84VRR33 pKa = 11.84SPRR36 pKa = 11.84QSVPLPKK43 pKa = 9.9KK44 pKa = 9.8RR45 pKa = 11.84LSPRR49 pKa = 11.84KK50 pKa = 8.85RR51 pKa = 11.84LLPRR55 pKa = 11.84SVLPRR60 pKa = 11.84KK61 pKa = 9.44RR62 pKa = 11.84RR63 pKa = 11.84PNQRR67 pKa = 11.84TSLEE71 pKa = 4.23KK72 pKa = 10.26ISPKK76 pKa = 10.23RR77 pKa = 11.84STPCPPIPTTKK88 pKa = 10.55LSLRR92 pKa = 11.84KK93 pKa = 9.43RR94 pKa = 11.84NPRR97 pKa = 11.84KK98 pKa = 9.74RR99 pKa = 11.84PLLRR103 pKa = 11.84RR104 pKa = 11.84SPPRR108 pKa = 11.84KK109 pKa = 9.38LLHH112 pKa = 5.7LRR114 pKa = 11.84LRR116 pKa = 11.84KK117 pKa = 9.29RR118 pKa = 11.84RR119 pKa = 11.84LSLRR123 pKa = 11.84RR124 pKa = 11.84KK125 pKa = 7.9QPQRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84MRR133 pKa = 11.84KK134 pKa = 9.18RR135 pKa = 11.84MSIWRR140 pKa = 11.84MPLSTKK146 pKa = 9.25RR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84VVASARR155 pKa = 11.84SLHH158 pKa = 6.75RR159 pKa = 11.84KK160 pKa = 8.59RR161 pKa = 11.84QLSLPRR167 pKa = 11.84RR168 pKa = 11.84KK169 pKa = 9.64PNPPPRR175 pKa = 11.84QRR177 pKa = 11.84RR178 pKa = 11.84ARR180 pKa = 11.84KK181 pKa = 8.7LLRR184 pKa = 11.84MMEE187 pKa = 4.3TMLMTDD193 pKa = 3.96DD194 pKa = 4.3YY195 pKa = 11.31LPVVPLQCFISFDD208 pKa = 3.42
Molecular weight: 25.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5698666 |
49 |
5063 |
401.9 |
44.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.089 ± 0.019 | 1.256 ± 0.008 |
5.31 ± 0.014 | 5.759 ± 0.021 |
3.867 ± 0.014 | 6.378 ± 0.02 |
2.547 ± 0.009 | 5.424 ± 0.015 |
4.719 ± 0.021 | 9.334 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.141 ± 0.008 | 3.725 ± 0.012 |
6.335 ± 0.03 | 3.706 ± 0.015 |
5.733 ± 0.018 | 9.033 ± 0.026 |
6.212 ± 0.015 | 6.21 ± 0.015 |
1.433 ± 0.008 | 2.786 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
