
Ferrovum sp. Z-31
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Ferrovales; Ferrovaceae; Ferrovum; unclassified Ferrovum
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
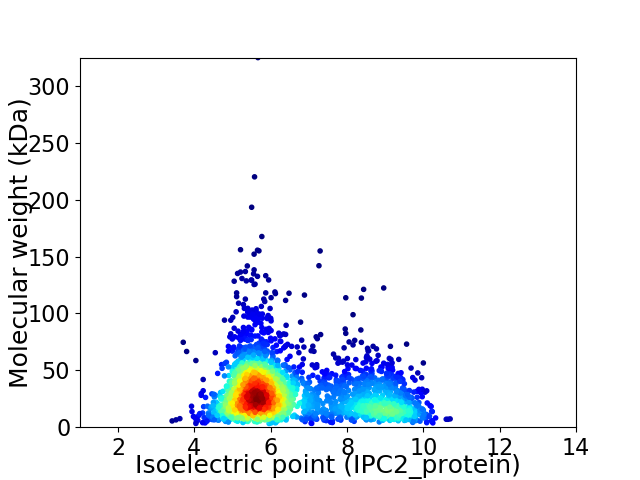
Virtual 2D-PAGE plot for 2451 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A149VYT0|A0A149VYT0_9PROT Probable Fe(2+)-trafficking protein OS=Ferrovum sp. Z-31 OX=1789004 GN=FEMY_11090 PE=3 SV=1
MM1 pKa = 7.64SINMSTQGVEE11 pKa = 3.53IARR14 pKa = 11.84YY15 pKa = 8.5AGAMYY20 pKa = 10.89GLVLDD25 pKa = 4.82DD26 pKa = 3.74ATVVSVEE33 pKa = 3.83NAANAGGSSLNAVMNQVYY51 pKa = 9.87AADD54 pKa = 4.48FSSISNATVATTVVTNLGLTGSLQSQAQAYY84 pKa = 9.69VLAQLNAAPAGSQGATIMTILNMFGQMTSDD114 pKa = 4.05PVWGAAATAWEE125 pKa = 4.39NKK127 pKa = 8.67VSEE130 pKa = 4.43SVTYY134 pKa = 9.73GQNKK138 pKa = 9.63ANVANSSIGGMSPTPVGGTYY158 pKa = 11.08DD159 pKa = 3.5LTTGVDD165 pKa = 3.69TLSGGPNATFIADD178 pKa = 3.52NTGTKK183 pKa = 8.91TLSAADD189 pKa = 4.32TIAATGTGNTLKK201 pKa = 10.74VYY203 pKa = 10.45LAAADD208 pKa = 3.86TTTGGTAGNITGVQNLYY225 pKa = 10.16INHH228 pKa = 7.1AGATAALTQDD238 pKa = 4.18FSTSSFTSITVDD250 pKa = 3.29SEE252 pKa = 4.65AFGAAALTLKK262 pKa = 10.54GQALTLEE269 pKa = 4.24NTGYY273 pKa = 10.9GATITDD279 pKa = 3.76TTDD282 pKa = 2.56TSLTVTVSAMSAGTLTTTGASKK304 pKa = 9.68ATTLNLVSSGTITGGNVVTLSTNAIDD330 pKa = 3.44TALNVSGATAITVTAGITGSADD352 pKa = 3.12LTSITDD358 pKa = 3.28TGTGGNTFDD367 pKa = 4.49ISTAIANAAFTFTGGSGGDD386 pKa = 3.69TLILAAGDD394 pKa = 3.91LTTLTSGSQLNGGGSASAPATLEE417 pKa = 3.98VNDD420 pKa = 4.52TSFSTAAYY428 pKa = 6.49TALNATTNFQILDD441 pKa = 3.77LNAAAGTTINASLITAGFHH460 pKa = 4.24NHH462 pKa = 5.51FAISAGSTNTISNMADD478 pKa = 2.89ASTVDD483 pKa = 3.25ISSAATSDD491 pKa = 3.5VLGGVVGAHH500 pKa = 5.47TLNLNLQSGAATMTEE515 pKa = 3.91GGITVTGLTTINLTSNTSTAGDD537 pKa = 4.0TNVVTAFVNSDD548 pKa = 2.76NTTFNVTGSAALTMAVAAATTTGDD572 pKa = 4.06TINASAFTGAFTLTATSGKK591 pKa = 10.49GDD593 pKa = 3.85IISTGSGTTSITDD606 pKa = 3.46TASATGNTDD615 pKa = 3.07TLLAGHH621 pKa = 6.3TAIDD625 pKa = 4.41TINTTANLPPAATTYY640 pKa = 9.1TATTLTAAMDD650 pKa = 3.76QISNFNIGATASDD663 pKa = 3.84ILKK666 pKa = 9.41MDD668 pKa = 4.09NGTKK672 pKa = 10.37AVGVSADD679 pKa = 4.48LGGTWTVTNGIATTSGTNTAAAFIAAVDD707 pKa = 3.86AATGTAGDD715 pKa = 3.75VVAYY719 pKa = 9.31TNGTNTYY726 pKa = 8.1VAAMDD731 pKa = 3.96GVVGKK736 pKa = 10.3AYY738 pKa = 10.11VVEE741 pKa = 4.03LVGVHH746 pKa = 5.78TATAVGITAAANTIHH761 pKa = 7.11IAA763 pKa = 3.36
MM1 pKa = 7.64SINMSTQGVEE11 pKa = 3.53IARR14 pKa = 11.84YY15 pKa = 8.5AGAMYY20 pKa = 10.89GLVLDD25 pKa = 4.82DD26 pKa = 3.74ATVVSVEE33 pKa = 3.83NAANAGGSSLNAVMNQVYY51 pKa = 9.87AADD54 pKa = 4.48FSSISNATVATTVVTNLGLTGSLQSQAQAYY84 pKa = 9.69VLAQLNAAPAGSQGATIMTILNMFGQMTSDD114 pKa = 4.05PVWGAAATAWEE125 pKa = 4.39NKK127 pKa = 8.67VSEE130 pKa = 4.43SVTYY134 pKa = 9.73GQNKK138 pKa = 9.63ANVANSSIGGMSPTPVGGTYY158 pKa = 11.08DD159 pKa = 3.5LTTGVDD165 pKa = 3.69TLSGGPNATFIADD178 pKa = 3.52NTGTKK183 pKa = 8.91TLSAADD189 pKa = 4.32TIAATGTGNTLKK201 pKa = 10.74VYY203 pKa = 10.45LAAADD208 pKa = 3.86TTTGGTAGNITGVQNLYY225 pKa = 10.16INHH228 pKa = 7.1AGATAALTQDD238 pKa = 4.18FSTSSFTSITVDD250 pKa = 3.29SEE252 pKa = 4.65AFGAAALTLKK262 pKa = 10.54GQALTLEE269 pKa = 4.24NTGYY273 pKa = 10.9GATITDD279 pKa = 3.76TTDD282 pKa = 2.56TSLTVTVSAMSAGTLTTTGASKK304 pKa = 9.68ATTLNLVSSGTITGGNVVTLSTNAIDD330 pKa = 3.44TALNVSGATAITVTAGITGSADD352 pKa = 3.12LTSITDD358 pKa = 3.28TGTGGNTFDD367 pKa = 4.49ISTAIANAAFTFTGGSGGDD386 pKa = 3.69TLILAAGDD394 pKa = 3.91LTTLTSGSQLNGGGSASAPATLEE417 pKa = 3.98VNDD420 pKa = 4.52TSFSTAAYY428 pKa = 6.49TALNATTNFQILDD441 pKa = 3.77LNAAAGTTINASLITAGFHH460 pKa = 4.24NHH462 pKa = 5.51FAISAGSTNTISNMADD478 pKa = 2.89ASTVDD483 pKa = 3.25ISSAATSDD491 pKa = 3.5VLGGVVGAHH500 pKa = 5.47TLNLNLQSGAATMTEE515 pKa = 3.91GGITVTGLTTINLTSNTSTAGDD537 pKa = 4.0TNVVTAFVNSDD548 pKa = 2.76NTTFNVTGSAALTMAVAAATTTGDD572 pKa = 4.06TINASAFTGAFTLTATSGKK591 pKa = 10.49GDD593 pKa = 3.85IISTGSGTTSITDD606 pKa = 3.46TASATGNTDD615 pKa = 3.07TLLAGHH621 pKa = 6.3TAIDD625 pKa = 4.41TINTTANLPPAATTYY640 pKa = 9.1TATTLTAAMDD650 pKa = 3.76QISNFNIGATASDD663 pKa = 3.84ILKK666 pKa = 9.41MDD668 pKa = 4.09NGTKK672 pKa = 10.37AVGVSADD679 pKa = 4.48LGGTWTVTNGIATTSGTNTAAAFIAAVDD707 pKa = 3.86AATGTAGDD715 pKa = 3.75VVAYY719 pKa = 9.31TNGTNTYY726 pKa = 8.1VAAMDD731 pKa = 3.96GVVGKK736 pKa = 10.3AYY738 pKa = 10.11VVEE741 pKa = 4.03LVGVHH746 pKa = 5.78TATAVGITAAANTIHH761 pKa = 7.11IAA763 pKa = 3.36
Molecular weight: 74.65 kDa
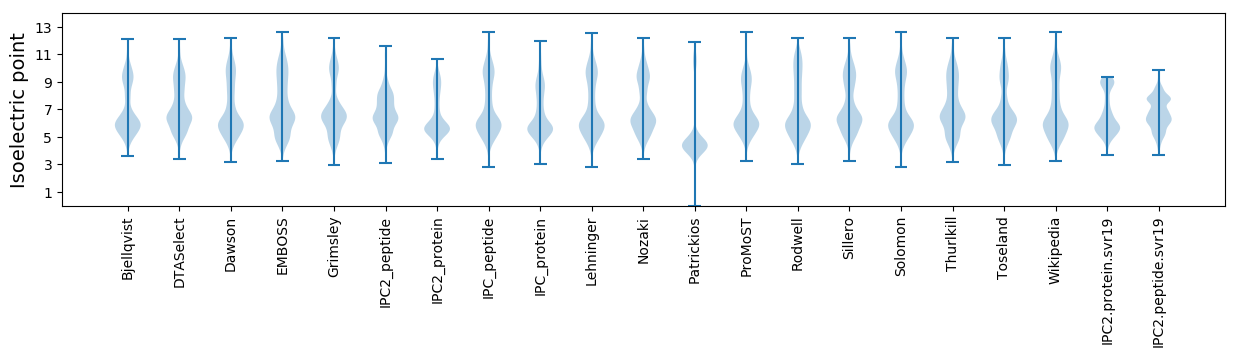
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A149VV54|A0A149VV54_9PROT Antitoxin DinJ OS=Ferrovum sp. Z-31 OX=1789004 GN=dinJ_2 PE=3 SV=1
MM1 pKa = 7.64ARR3 pKa = 11.84NCLPCSRR10 pKa = 11.84SMAQFPDD17 pKa = 3.42AEE19 pKa = 4.62TYY21 pKa = 10.68SPGEE25 pKa = 3.82IMGVCPIRR33 pKa = 11.84VTSSRR38 pKa = 11.84SPLTLMRR45 pKa = 11.84RR46 pKa = 11.84TQKK49 pKa = 10.47PFSALWKK56 pKa = 7.9VTRR59 pKa = 11.84STRR62 pKa = 11.84PARR65 pKa = 11.84RR66 pKa = 11.84SCSLVVSCGILGFSMGGVAEE86 pKa = 4.31KK87 pKa = 10.97YY88 pKa = 9.31EE89 pKa = 4.2RR90 pKa = 11.84TVSGAEE96 pKa = 3.68YY97 pKa = 10.8RR98 pKa = 11.84LNQWNNSSDD107 pKa = 3.47PQHH110 pKa = 6.45FLNFLPEE117 pKa = 4.14PHH119 pKa = 6.25GHH121 pKa = 6.08GSLRR125 pKa = 11.84PTLASLRR132 pKa = 11.84RR133 pKa = 11.84KK134 pKa = 9.99GCVSSSFEE142 pKa = 3.74VRR144 pKa = 11.84IFPIRR149 pKa = 11.84DD150 pKa = 3.29SIACMRR156 pKa = 11.84RR157 pKa = 11.84GSKK160 pKa = 10.09
MM1 pKa = 7.64ARR3 pKa = 11.84NCLPCSRR10 pKa = 11.84SMAQFPDD17 pKa = 3.42AEE19 pKa = 4.62TYY21 pKa = 10.68SPGEE25 pKa = 3.82IMGVCPIRR33 pKa = 11.84VTSSRR38 pKa = 11.84SPLTLMRR45 pKa = 11.84RR46 pKa = 11.84TQKK49 pKa = 10.47PFSALWKK56 pKa = 7.9VTRR59 pKa = 11.84STRR62 pKa = 11.84PARR65 pKa = 11.84RR66 pKa = 11.84SCSLVVSCGILGFSMGGVAEE86 pKa = 4.31KK87 pKa = 10.97YY88 pKa = 9.31EE89 pKa = 4.2RR90 pKa = 11.84TVSGAEE96 pKa = 3.68YY97 pKa = 10.8RR98 pKa = 11.84LNQWNNSSDD107 pKa = 3.47PQHH110 pKa = 6.45FLNFLPEE117 pKa = 4.14PHH119 pKa = 6.25GHH121 pKa = 6.08GSLRR125 pKa = 11.84PTLASLRR132 pKa = 11.84RR133 pKa = 11.84KK134 pKa = 9.99GCVSSSFEE142 pKa = 3.74VRR144 pKa = 11.84IFPIRR149 pKa = 11.84DD150 pKa = 3.29SIACMRR156 pKa = 11.84RR157 pKa = 11.84GSKK160 pKa = 10.09
Molecular weight: 17.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
734706 |
29 |
2891 |
299.8 |
33.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.455 ± 0.051 | 1.003 ± 0.018 |
4.949 ± 0.034 | 6.022 ± 0.051 |
3.795 ± 0.031 | 7.759 ± 0.053 |
2.772 ± 0.025 | 5.065 ± 0.033 |
3.544 ± 0.04 | 11.562 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.44 ± 0.023 | 2.996 ± 0.036 |
5.124 ± 0.031 | 4.518 ± 0.033 |
6.642 ± 0.047 | 5.801 ± 0.037 |
5.422 ± 0.037 | 7.158 ± 0.046 |
1.548 ± 0.024 | 2.425 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
