
Halomonas muralis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Halomonas
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
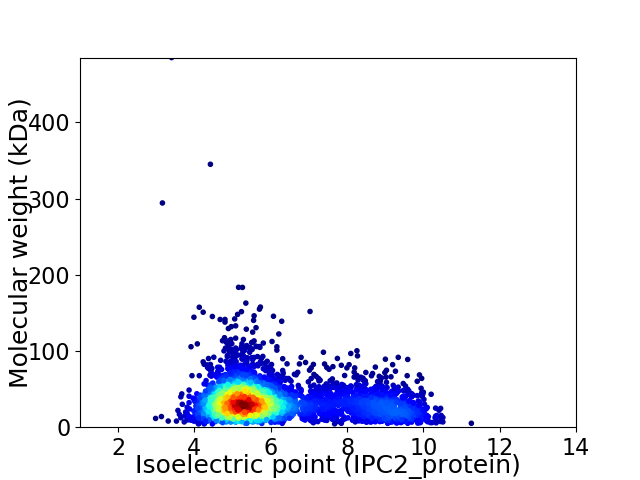
Virtual 2D-PAGE plot for 3825 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
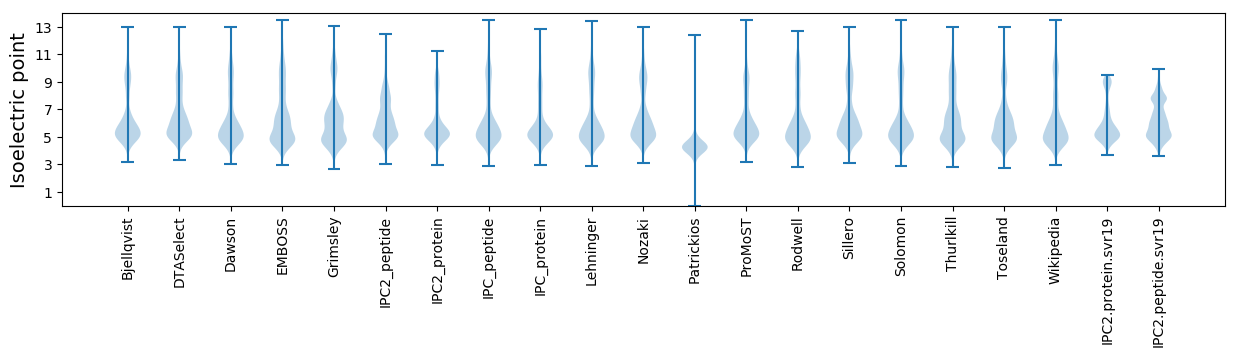
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9RWQ2|A0A1G9RWQ2_9GAMM N-succinylarginine dihydrolase OS=Halomonas muralis OX=119000 GN=astB PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGALGASAAAQAATVYY26 pKa = 10.54NQDD29 pKa = 2.95GTQLDD34 pKa = 3.83IYY36 pKa = 11.21GNIQIAYY43 pKa = 8.61YY44 pKa = 10.62SLDD47 pKa = 3.71AGDD50 pKa = 4.19EE51 pKa = 4.42SVDD54 pKa = 3.92EE55 pKa = 4.38IADD58 pKa = 3.64NGSTFGFAAQHH69 pKa = 5.85VINDD73 pKa = 3.85GLTGYY78 pKa = 10.81LKK80 pKa = 11.2LEE82 pKa = 4.31FDD84 pKa = 4.44DD85 pKa = 6.14FKK87 pKa = 11.61ADD89 pKa = 4.07EE90 pKa = 4.27IKK92 pKa = 11.09VRR94 pKa = 11.84GRR96 pKa = 11.84DD97 pKa = 3.36AGDD100 pKa = 3.13QAYY103 pKa = 10.69VGLKK107 pKa = 10.76GNFGDD112 pKa = 5.01ARR114 pKa = 11.84LGSYY118 pKa = 10.69DD119 pKa = 4.28PLIDD123 pKa = 5.54DD124 pKa = 5.75WIQDD128 pKa = 4.02PITNNEE134 pKa = 3.86FADD137 pKa = 3.85VSDD140 pKa = 3.95SSGLIVDD147 pKa = 4.63IYY149 pKa = 11.13GADD152 pKa = 3.57DD153 pKa = 3.83RR154 pKa = 11.84EE155 pKa = 4.24GDD157 pKa = 3.43KK158 pKa = 10.87FQYY161 pKa = 9.55TSPSFGGLQFAVGTQYY177 pKa = 11.35KK178 pKa = 10.6GDD180 pKa = 3.96AEE182 pKa = 4.23EE183 pKa = 4.34KK184 pKa = 10.34NAFIEE189 pKa = 4.3ADD191 pKa = 3.19GRR193 pKa = 11.84ASFFSGVKK201 pKa = 7.36YY202 pKa = 9.14TVGAFSIAGVYY213 pKa = 10.57DD214 pKa = 3.36NLAIYY219 pKa = 10.54DD220 pKa = 4.12GEE222 pKa = 4.56FNTAVDD228 pKa = 3.51VDD230 pKa = 4.0GDD232 pKa = 3.96GTADD236 pKa = 3.1IAAGDD241 pKa = 3.99DD242 pKa = 4.3FEE244 pKa = 7.6AGDD247 pKa = 3.79QYY249 pKa = 11.83GVTAQYY255 pKa = 11.28TMGALRR261 pKa = 11.84VALKK265 pKa = 9.21YY266 pKa = 10.29EE267 pKa = 4.3RR268 pKa = 11.84YY269 pKa = 8.47EE270 pKa = 4.33SEE272 pKa = 4.45VEE274 pKa = 4.5DD275 pKa = 3.73VQPDD279 pKa = 3.52VNFYY283 pKa = 11.25GIGARR288 pKa = 11.84YY289 pKa = 8.88GYY291 pKa = 11.51GMGDD295 pKa = 2.98IYY297 pKa = 11.28AAYY300 pKa = 9.82QYY302 pKa = 11.46VDD304 pKa = 2.88IGGSDD309 pKa = 3.65LEE311 pKa = 4.34EE312 pKa = 4.44TIEE315 pKa = 4.85EE316 pKa = 4.08DD317 pKa = 5.0DD318 pKa = 4.22EE319 pKa = 5.55NIGEE323 pKa = 4.3NDD325 pKa = 3.48DD326 pKa = 3.76SRR328 pKa = 11.84NEE330 pKa = 3.77ILVGVTYY337 pKa = 10.63NVSPAMYY344 pKa = 8.43VWLEE348 pKa = 3.74GGVRR352 pKa = 11.84DD353 pKa = 5.2QEE355 pKa = 4.9DD356 pKa = 3.7DD357 pKa = 4.33LGDD360 pKa = 4.05FTATGVTYY368 pKa = 10.81SFF370 pKa = 4.48
MM1 pKa = 7.71KK2 pKa = 9.06KK3 pKa = 8.27TLLATAIAGALGASAAAQAATVYY26 pKa = 10.54NQDD29 pKa = 2.95GTQLDD34 pKa = 3.83IYY36 pKa = 11.21GNIQIAYY43 pKa = 8.61YY44 pKa = 10.62SLDD47 pKa = 3.71AGDD50 pKa = 4.19EE51 pKa = 4.42SVDD54 pKa = 3.92EE55 pKa = 4.38IADD58 pKa = 3.64NGSTFGFAAQHH69 pKa = 5.85VINDD73 pKa = 3.85GLTGYY78 pKa = 10.81LKK80 pKa = 11.2LEE82 pKa = 4.31FDD84 pKa = 4.44DD85 pKa = 6.14FKK87 pKa = 11.61ADD89 pKa = 4.07EE90 pKa = 4.27IKK92 pKa = 11.09VRR94 pKa = 11.84GRR96 pKa = 11.84DD97 pKa = 3.36AGDD100 pKa = 3.13QAYY103 pKa = 10.69VGLKK107 pKa = 10.76GNFGDD112 pKa = 5.01ARR114 pKa = 11.84LGSYY118 pKa = 10.69DD119 pKa = 4.28PLIDD123 pKa = 5.54DD124 pKa = 5.75WIQDD128 pKa = 4.02PITNNEE134 pKa = 3.86FADD137 pKa = 3.85VSDD140 pKa = 3.95SSGLIVDD147 pKa = 4.63IYY149 pKa = 11.13GADD152 pKa = 3.57DD153 pKa = 3.83RR154 pKa = 11.84EE155 pKa = 4.24GDD157 pKa = 3.43KK158 pKa = 10.87FQYY161 pKa = 9.55TSPSFGGLQFAVGTQYY177 pKa = 11.35KK178 pKa = 10.6GDD180 pKa = 3.96AEE182 pKa = 4.23EE183 pKa = 4.34KK184 pKa = 10.34NAFIEE189 pKa = 4.3ADD191 pKa = 3.19GRR193 pKa = 11.84ASFFSGVKK201 pKa = 7.36YY202 pKa = 9.14TVGAFSIAGVYY213 pKa = 10.57DD214 pKa = 3.36NLAIYY219 pKa = 10.54DD220 pKa = 4.12GEE222 pKa = 4.56FNTAVDD228 pKa = 3.51VDD230 pKa = 4.0GDD232 pKa = 3.96GTADD236 pKa = 3.1IAAGDD241 pKa = 3.99DD242 pKa = 4.3FEE244 pKa = 7.6AGDD247 pKa = 3.79QYY249 pKa = 11.83GVTAQYY255 pKa = 11.28TMGALRR261 pKa = 11.84VALKK265 pKa = 9.21YY266 pKa = 10.29EE267 pKa = 4.3RR268 pKa = 11.84YY269 pKa = 8.47EE270 pKa = 4.33SEE272 pKa = 4.45VEE274 pKa = 4.5DD275 pKa = 3.73VQPDD279 pKa = 3.52VNFYY283 pKa = 11.25GIGARR288 pKa = 11.84YY289 pKa = 8.88GYY291 pKa = 11.51GMGDD295 pKa = 2.98IYY297 pKa = 11.28AAYY300 pKa = 9.82QYY302 pKa = 11.46VDD304 pKa = 2.88IGGSDD309 pKa = 3.65LEE311 pKa = 4.34EE312 pKa = 4.44TIEE315 pKa = 4.85EE316 pKa = 4.08DD317 pKa = 5.0DD318 pKa = 4.22EE319 pKa = 5.55NIGEE323 pKa = 4.3NDD325 pKa = 3.48DD326 pKa = 3.76SRR328 pKa = 11.84NEE330 pKa = 3.77ILVGVTYY337 pKa = 10.63NVSPAMYY344 pKa = 8.43VWLEE348 pKa = 3.74GGVRR352 pKa = 11.84DD353 pKa = 5.2QEE355 pKa = 4.9DD356 pKa = 3.7DD357 pKa = 4.33LGDD360 pKa = 4.05FTATGVTYY368 pKa = 10.81SFF370 pKa = 4.48
Molecular weight: 39.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9L1K9|A0A1G9L1K9_9GAMM Nucleobase:cation symporter-1 NCS1 family OS=Halomonas muralis OX=119000 GN=SAMN05661010_01983 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.34RR12 pKa = 11.84KK13 pKa = 9.1RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1244899 |
39 |
4634 |
325.5 |
35.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.315 ± 0.051 | 0.97 ± 0.014 |
5.822 ± 0.038 | 6.247 ± 0.036 |
3.52 ± 0.028 | 8.273 ± 0.04 |
2.417 ± 0.02 | 4.972 ± 0.029 |
2.691 ± 0.031 | 11.36 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.479 ± 0.018 | 2.631 ± 0.025 |
4.89 ± 0.026 | 3.772 ± 0.029 |
7.179 ± 0.051 | 5.474 ± 0.025 |
4.988 ± 0.035 | 7.115 ± 0.03 |
1.468 ± 0.016 | 2.417 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
