
Hubei diptera virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
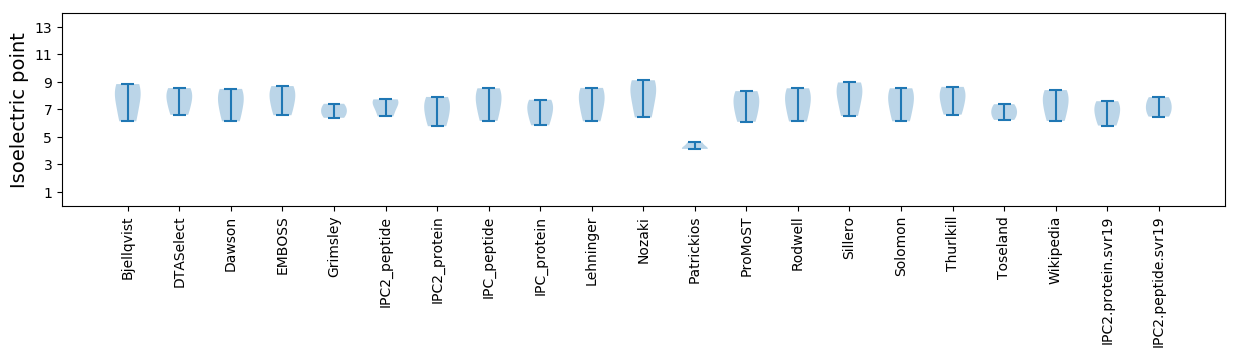
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KEK9|A0A1L3KEK9_9VIRU C2H2-type domain-containing protein OS=Hubei diptera virus 12 OX=1922873 PE=4 SV=1
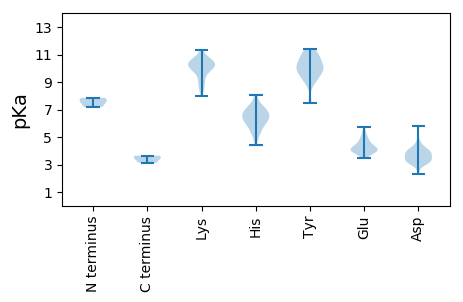
MM1 pKa = 7.62EE2 pKa = 5.47ARR4 pKa = 11.84NCFSSLSWTIPEE16 pKa = 4.33DD17 pKa = 3.52FCTFTHH23 pKa = 6.3YY24 pKa = 10.71LRR26 pKa = 11.84ALDD29 pKa = 3.99RR30 pKa = 11.84LEE32 pKa = 4.37MSSSPGYY39 pKa = 9.71PYY41 pKa = 9.89MRR43 pKa = 11.84RR44 pKa = 11.84APTNRR49 pKa = 11.84DD50 pKa = 3.02LFKK53 pKa = 11.06VDD55 pKa = 4.69DD56 pKa = 4.29VGNKK60 pKa = 9.91NSDD63 pKa = 3.27VVLIFWDD70 pKa = 3.38IVRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84LEE77 pKa = 4.0EE78 pKa = 4.93LGDD81 pKa = 4.04ADD83 pKa = 5.77PIRR86 pKa = 11.84LFIKK90 pKa = 10.29QEE92 pKa = 3.51PHH94 pKa = 5.41KK95 pKa = 10.53QKK97 pKa = 10.98KK98 pKa = 10.03LDD100 pKa = 3.87DD101 pKa = 3.61GRR103 pKa = 11.84FRR105 pKa = 11.84LISSVSVVDD114 pKa = 4.52QIIDD118 pKa = 3.3HH119 pKa = 6.08MLFAEE124 pKa = 5.01MNDD127 pKa = 2.98QMTQNWIRR135 pKa = 11.84NPIKK139 pKa = 10.57IGWTPVKK146 pKa = 10.3GGWRR150 pKa = 11.84AFPPGKK156 pKa = 9.75RR157 pKa = 11.84VAVDD161 pKa = 3.24KK162 pKa = 10.77SHH164 pKa = 6.98WDD166 pKa = 3.2WTVSLWMLDD175 pKa = 3.04CVLQLRR181 pKa = 11.84GEE183 pKa = 4.32LCVTRR188 pKa = 11.84GEE190 pKa = 5.66LFDD193 pKa = 4.3KK194 pKa = 10.46WLLLAKK200 pKa = 10.06YY201 pKa = 9.51RR202 pKa = 11.84YY203 pKa = 9.63SEE205 pKa = 4.79LFDD208 pKa = 4.08GPLFITSGGVLLRR221 pKa = 11.84QKK223 pKa = 10.75VPGVQKK229 pKa = 10.45SGCVNTLADD238 pKa = 3.89NSLMQWILHH247 pKa = 5.26ARR249 pKa = 11.84VMLEE253 pKa = 3.86HH254 pKa = 7.01GLNPHH259 pKa = 6.81DD260 pKa = 3.53VDD262 pKa = 3.48MWVMGDD268 pKa = 3.99DD269 pKa = 3.97TSQSAIVKK277 pKa = 10.1DD278 pKa = 3.96YY279 pKa = 11.05IDD281 pKa = 3.72WLGEE285 pKa = 3.88YY286 pKa = 10.08CVVKK290 pKa = 10.36EE291 pKa = 4.07SQIAPEE297 pKa = 3.89FCGFRR302 pKa = 11.84FMEE305 pKa = 4.74AGRR308 pKa = 11.84VEE310 pKa = 4.01PLYY313 pKa = 10.61KK314 pKa = 10.41GKK316 pKa = 9.54HH317 pKa = 5.09AFNLLHH323 pKa = 6.81MDD325 pKa = 4.22DD326 pKa = 5.64KK327 pKa = 11.32YY328 pKa = 11.38ATDD331 pKa = 3.37IANSYY336 pKa = 9.83SLLYY340 pKa = 10.3HH341 pKa = 7.17RR342 pKa = 11.84STHH345 pKa = 6.11RR346 pKa = 11.84DD347 pKa = 2.32WMRR350 pKa = 11.84NLFISMGINIYY361 pKa = 10.05PSYY364 pKa = 10.85YY365 pKa = 10.35LDD367 pKa = 3.86TIYY370 pKa = 11.21DD371 pKa = 3.73GVEE374 pKa = 3.49
MM1 pKa = 7.62EE2 pKa = 5.47ARR4 pKa = 11.84NCFSSLSWTIPEE16 pKa = 4.33DD17 pKa = 3.52FCTFTHH23 pKa = 6.3YY24 pKa = 10.71LRR26 pKa = 11.84ALDD29 pKa = 3.99RR30 pKa = 11.84LEE32 pKa = 4.37MSSSPGYY39 pKa = 9.71PYY41 pKa = 9.89MRR43 pKa = 11.84RR44 pKa = 11.84APTNRR49 pKa = 11.84DD50 pKa = 3.02LFKK53 pKa = 11.06VDD55 pKa = 4.69DD56 pKa = 4.29VGNKK60 pKa = 9.91NSDD63 pKa = 3.27VVLIFWDD70 pKa = 3.38IVRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84LEE77 pKa = 4.0EE78 pKa = 4.93LGDD81 pKa = 4.04ADD83 pKa = 5.77PIRR86 pKa = 11.84LFIKK90 pKa = 10.29QEE92 pKa = 3.51PHH94 pKa = 5.41KK95 pKa = 10.53QKK97 pKa = 10.98KK98 pKa = 10.03LDD100 pKa = 3.87DD101 pKa = 3.61GRR103 pKa = 11.84FRR105 pKa = 11.84LISSVSVVDD114 pKa = 4.52QIIDD118 pKa = 3.3HH119 pKa = 6.08MLFAEE124 pKa = 5.01MNDD127 pKa = 2.98QMTQNWIRR135 pKa = 11.84NPIKK139 pKa = 10.57IGWTPVKK146 pKa = 10.3GGWRR150 pKa = 11.84AFPPGKK156 pKa = 9.75RR157 pKa = 11.84VAVDD161 pKa = 3.24KK162 pKa = 10.77SHH164 pKa = 6.98WDD166 pKa = 3.2WTVSLWMLDD175 pKa = 3.04CVLQLRR181 pKa = 11.84GEE183 pKa = 4.32LCVTRR188 pKa = 11.84GEE190 pKa = 5.66LFDD193 pKa = 4.3KK194 pKa = 10.46WLLLAKK200 pKa = 10.06YY201 pKa = 9.51RR202 pKa = 11.84YY203 pKa = 9.63SEE205 pKa = 4.79LFDD208 pKa = 4.08GPLFITSGGVLLRR221 pKa = 11.84QKK223 pKa = 10.75VPGVQKK229 pKa = 10.45SGCVNTLADD238 pKa = 3.89NSLMQWILHH247 pKa = 5.26ARR249 pKa = 11.84VMLEE253 pKa = 3.86HH254 pKa = 7.01GLNPHH259 pKa = 6.81DD260 pKa = 3.53VDD262 pKa = 3.48MWVMGDD268 pKa = 3.99DD269 pKa = 3.97TSQSAIVKK277 pKa = 10.1DD278 pKa = 3.96YY279 pKa = 11.05IDD281 pKa = 3.72WLGEE285 pKa = 3.88YY286 pKa = 10.08CVVKK290 pKa = 10.36EE291 pKa = 4.07SQIAPEE297 pKa = 3.89FCGFRR302 pKa = 11.84FMEE305 pKa = 4.74AGRR308 pKa = 11.84VEE310 pKa = 4.01PLYY313 pKa = 10.61KK314 pKa = 10.41GKK316 pKa = 9.54HH317 pKa = 5.09AFNLLHH323 pKa = 6.81MDD325 pKa = 4.22DD326 pKa = 5.64KK327 pKa = 11.32YY328 pKa = 11.38ATDD331 pKa = 3.37IANSYY336 pKa = 9.83SLLYY340 pKa = 10.3HH341 pKa = 7.17RR342 pKa = 11.84STHH345 pKa = 6.11RR346 pKa = 11.84DD347 pKa = 2.32WMRR350 pKa = 11.84NLFISMGINIYY361 pKa = 10.05PSYY364 pKa = 10.85YY365 pKa = 10.35LDD367 pKa = 3.86TIYY370 pKa = 11.21DD371 pKa = 3.73GVEE374 pKa = 3.49
Molecular weight: 43.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KER4|A0A1L3KER4_9VIRU C2H2-type domain-containing protein OS=Hubei diptera virus 12 OX=1922873 PE=4 SV=1
MM1 pKa = 7.85DD2 pKa = 3.83TQNNDD7 pKa = 2.36THH9 pKa = 6.97RR10 pKa = 11.84RR11 pKa = 11.84QTLGPTNVVVGWILDD26 pKa = 3.76IFWPLIPVFKK36 pKa = 9.91MLNVVCKK43 pKa = 9.96VCVVYY48 pKa = 10.75ALAVAFTGVYY58 pKa = 10.23RR59 pKa = 11.84LVLWISRR66 pKa = 11.84TPAQVTMNVTVFVTKK81 pKa = 10.63LFHH84 pKa = 6.98RR85 pKa = 11.84LVLLVHH91 pKa = 6.22WPIEE95 pKa = 4.05KK96 pKa = 10.21LVEE99 pKa = 4.01LMFMEE104 pKa = 5.31PEE106 pKa = 4.34PEE108 pKa = 5.14PIPLTWTEE116 pKa = 3.92TLLEE120 pKa = 4.1GLEE123 pKa = 4.19TKK125 pKa = 9.82IRR127 pKa = 11.84SILPVVAMITQSVVVYY143 pKa = 10.06IVGSFIASYY152 pKa = 10.99LLYY155 pKa = 10.53RR156 pKa = 11.84LTKK159 pKa = 10.31KK160 pKa = 9.84PVKK163 pKa = 8.69TLVMRR168 pKa = 11.84SRR170 pKa = 11.84GVYY173 pKa = 9.01VGEE176 pKa = 4.02AMRR179 pKa = 11.84EE180 pKa = 4.0GSQFVPVKK188 pKa = 10.36SIPPGQVSIMKK199 pKa = 10.29SGLLTDD205 pKa = 3.03SHH207 pKa = 8.06IGYY210 pKa = 9.84GLRR213 pKa = 11.84VHH215 pKa = 7.28DD216 pKa = 4.61CLVTPAHH223 pKa = 6.03VADD226 pKa = 4.2LEE228 pKa = 4.55KK229 pKa = 10.59TFVLANRR236 pKa = 11.84GVKK239 pKa = 10.05VLFTPTTRR247 pKa = 11.84IPSRR251 pKa = 11.84VIPDD255 pKa = 3.74LVYY258 pKa = 10.56IPVEE262 pKa = 3.65PSIWARR268 pKa = 11.84LGATAVKK275 pKa = 8.6MQRR278 pKa = 11.84GVPKK282 pKa = 10.33LANQVTCTGLPGATTGLLRR301 pKa = 11.84STTRR305 pKa = 11.84MGVYY309 pKa = 9.81SYY311 pKa = 11.14SGSTLPGMSGAGYY324 pKa = 9.88FVGNCCFGMHH334 pKa = 6.89TGNIAGEE341 pKa = 4.23NVGIASSLIMRR352 pKa = 11.84EE353 pKa = 3.6WRR355 pKa = 11.84CIYY358 pKa = 10.29KK359 pKa = 10.68GEE361 pKa = 4.11SSPDD365 pKa = 3.49FVDD368 pKa = 3.37TSVPIVKK375 pKa = 8.68KK376 pKa = 8.56TWDD379 pKa = 3.7DD380 pKa = 3.87VEE382 pKa = 4.73IDD384 pKa = 3.6AAATKK389 pKa = 10.3AWLEE393 pKa = 4.88DD394 pKa = 3.25EE395 pKa = 5.74DD396 pKa = 4.02FWEE399 pKa = 5.13DD400 pKa = 4.22RR401 pKa = 11.84VANWGQKK408 pKa = 9.61SFAEE412 pKa = 3.95NAFITEE418 pKa = 4.18EE419 pKa = 4.3TVVKK423 pKa = 10.47QQGNDD428 pKa = 3.69GEE430 pKa = 4.45EE431 pKa = 3.73KK432 pKa = 10.25VYY434 pKa = 9.77RR435 pKa = 11.84TTVVPSDD442 pKa = 3.24LTQRR446 pKa = 11.84VSLLEE451 pKa = 3.65QAMAKK456 pKa = 10.09VGLVKK461 pKa = 10.41CAQCDD466 pKa = 3.42EE467 pKa = 4.45VFVGKK472 pKa = 7.99TLEE475 pKa = 3.96QHH477 pKa = 6.48KK478 pKa = 10.05KK479 pKa = 9.3DD480 pKa = 3.22HH481 pKa = 6.44AVRR484 pKa = 11.84HH485 pKa = 5.76PCEE488 pKa = 3.87QCDD491 pKa = 3.93MVCLTVEE498 pKa = 3.86KK499 pKa = 10.49LSRR502 pKa = 11.84HH503 pKa = 4.41VKK505 pKa = 8.75EE506 pKa = 3.6FHH508 pKa = 7.14SRR510 pKa = 11.84VEE512 pKa = 4.16CDD514 pKa = 2.84RR515 pKa = 11.84CSTVCKK521 pKa = 10.88NMDD524 pKa = 3.16KK525 pKa = 10.74LANHH529 pKa = 7.35RR530 pKa = 11.84KK531 pKa = 8.13NCKK534 pKa = 9.87AVGDD538 pKa = 4.08VQLMEE543 pKa = 4.54VPSSSTHH550 pKa = 5.6PRR552 pKa = 11.84TPLVEE557 pKa = 3.98THH559 pKa = 6.41PQVPVAEE566 pKa = 4.36EE567 pKa = 4.03VKK569 pKa = 10.42EE570 pKa = 4.08SAFAIDD576 pKa = 3.53SRR578 pKa = 11.84KK579 pKa = 9.55LVKK582 pKa = 10.61NSPFLGQRR590 pKa = 11.84QPSRR594 pKa = 11.84TKK596 pKa = 10.06NSKK599 pKa = 9.79RR600 pKa = 11.84SQSGSSLKK608 pKa = 10.42VASPHH613 pKa = 6.41SPSQGEE619 pKa = 4.24MLSQILTSQKK629 pKa = 10.91SMQEE633 pKa = 3.67SFEE636 pKa = 4.65RR637 pKa = 11.84FLQVMVGQVSVTKK650 pKa = 10.76QNN652 pKa = 3.12
MM1 pKa = 7.85DD2 pKa = 3.83TQNNDD7 pKa = 2.36THH9 pKa = 6.97RR10 pKa = 11.84RR11 pKa = 11.84QTLGPTNVVVGWILDD26 pKa = 3.76IFWPLIPVFKK36 pKa = 9.91MLNVVCKK43 pKa = 9.96VCVVYY48 pKa = 10.75ALAVAFTGVYY58 pKa = 10.23RR59 pKa = 11.84LVLWISRR66 pKa = 11.84TPAQVTMNVTVFVTKK81 pKa = 10.63LFHH84 pKa = 6.98RR85 pKa = 11.84LVLLVHH91 pKa = 6.22WPIEE95 pKa = 4.05KK96 pKa = 10.21LVEE99 pKa = 4.01LMFMEE104 pKa = 5.31PEE106 pKa = 4.34PEE108 pKa = 5.14PIPLTWTEE116 pKa = 3.92TLLEE120 pKa = 4.1GLEE123 pKa = 4.19TKK125 pKa = 9.82IRR127 pKa = 11.84SILPVVAMITQSVVVYY143 pKa = 10.06IVGSFIASYY152 pKa = 10.99LLYY155 pKa = 10.53RR156 pKa = 11.84LTKK159 pKa = 10.31KK160 pKa = 9.84PVKK163 pKa = 8.69TLVMRR168 pKa = 11.84SRR170 pKa = 11.84GVYY173 pKa = 9.01VGEE176 pKa = 4.02AMRR179 pKa = 11.84EE180 pKa = 4.0GSQFVPVKK188 pKa = 10.36SIPPGQVSIMKK199 pKa = 10.29SGLLTDD205 pKa = 3.03SHH207 pKa = 8.06IGYY210 pKa = 9.84GLRR213 pKa = 11.84VHH215 pKa = 7.28DD216 pKa = 4.61CLVTPAHH223 pKa = 6.03VADD226 pKa = 4.2LEE228 pKa = 4.55KK229 pKa = 10.59TFVLANRR236 pKa = 11.84GVKK239 pKa = 10.05VLFTPTTRR247 pKa = 11.84IPSRR251 pKa = 11.84VIPDD255 pKa = 3.74LVYY258 pKa = 10.56IPVEE262 pKa = 3.65PSIWARR268 pKa = 11.84LGATAVKK275 pKa = 8.6MQRR278 pKa = 11.84GVPKK282 pKa = 10.33LANQVTCTGLPGATTGLLRR301 pKa = 11.84STTRR305 pKa = 11.84MGVYY309 pKa = 9.81SYY311 pKa = 11.14SGSTLPGMSGAGYY324 pKa = 9.88FVGNCCFGMHH334 pKa = 6.89TGNIAGEE341 pKa = 4.23NVGIASSLIMRR352 pKa = 11.84EE353 pKa = 3.6WRR355 pKa = 11.84CIYY358 pKa = 10.29KK359 pKa = 10.68GEE361 pKa = 4.11SSPDD365 pKa = 3.49FVDD368 pKa = 3.37TSVPIVKK375 pKa = 8.68KK376 pKa = 8.56TWDD379 pKa = 3.7DD380 pKa = 3.87VEE382 pKa = 4.73IDD384 pKa = 3.6AAATKK389 pKa = 10.3AWLEE393 pKa = 4.88DD394 pKa = 3.25EE395 pKa = 5.74DD396 pKa = 4.02FWEE399 pKa = 5.13DD400 pKa = 4.22RR401 pKa = 11.84VANWGQKK408 pKa = 9.61SFAEE412 pKa = 3.95NAFITEE418 pKa = 4.18EE419 pKa = 4.3TVVKK423 pKa = 10.47QQGNDD428 pKa = 3.69GEE430 pKa = 4.45EE431 pKa = 3.73KK432 pKa = 10.25VYY434 pKa = 9.77RR435 pKa = 11.84TTVVPSDD442 pKa = 3.24LTQRR446 pKa = 11.84VSLLEE451 pKa = 3.65QAMAKK456 pKa = 10.09VGLVKK461 pKa = 10.41CAQCDD466 pKa = 3.42EE467 pKa = 4.45VFVGKK472 pKa = 7.99TLEE475 pKa = 3.96QHH477 pKa = 6.48KK478 pKa = 10.05KK479 pKa = 9.3DD480 pKa = 3.22HH481 pKa = 6.44AVRR484 pKa = 11.84HH485 pKa = 5.76PCEE488 pKa = 3.87QCDD491 pKa = 3.93MVCLTVEE498 pKa = 3.86KK499 pKa = 10.49LSRR502 pKa = 11.84HH503 pKa = 4.41VKK505 pKa = 8.75EE506 pKa = 3.6FHH508 pKa = 7.14SRR510 pKa = 11.84VEE512 pKa = 4.16CDD514 pKa = 2.84RR515 pKa = 11.84CSTVCKK521 pKa = 10.88NMDD524 pKa = 3.16KK525 pKa = 10.74LANHH529 pKa = 7.35RR530 pKa = 11.84KK531 pKa = 8.13NCKK534 pKa = 9.87AVGDD538 pKa = 4.08VQLMEE543 pKa = 4.54VPSSSTHH550 pKa = 5.6PRR552 pKa = 11.84TPLVEE557 pKa = 3.98THH559 pKa = 6.41PQVPVAEE566 pKa = 4.36EE567 pKa = 4.03VKK569 pKa = 10.42EE570 pKa = 4.08SAFAIDD576 pKa = 3.53SRR578 pKa = 11.84KK579 pKa = 9.55LVKK582 pKa = 10.61NSPFLGQRR590 pKa = 11.84QPSRR594 pKa = 11.84TKK596 pKa = 10.06NSKK599 pKa = 9.79RR600 pKa = 11.84SQSGSSLKK608 pKa = 10.42VASPHH613 pKa = 6.41SPSQGEE619 pKa = 4.24MLSQILTSQKK629 pKa = 10.91SMQEE633 pKa = 3.67SFEE636 pKa = 4.65RR637 pKa = 11.84FLQVMVGQVSVTKK650 pKa = 10.76QNN652 pKa = 3.12
Molecular weight: 72.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1488 |
374 |
652 |
496.0 |
55.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.847 ± 0.82 | 2.352 ± 0.16 |
5.511 ± 1.09 | 4.839 ± 0.661 |
3.427 ± 0.418 | 6.317 ± 0.254 |
2.755 ± 0.16 | 4.503 ± 0.358 |
5.04 ± 0.698 | 8.602 ± 0.52 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.957 ± 0.523 | 3.091 ± 0.212 |
5.04 ± 0.28 | 4.234 ± 0.483 |
5.847 ± 0.343 | 7.93 ± 0.768 |
6.855 ± 1.065 | 10.013 ± 1.295 |
2.083 ± 0.451 | 2.755 ± 0.451 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
