
Methanospirillum lacunae
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanospirillaceae; Methanospirillum
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
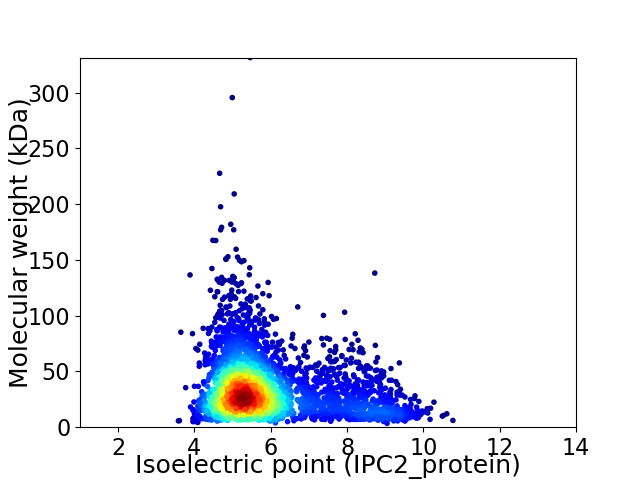
Virtual 2D-PAGE plot for 3377 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V2NFG1|A0A2V2NFG1_9EURY Methyl-accepting chemotaxis protein OS=Methanospirillum lacunae OX=668570 GN=DK846_04065 PE=4 SV=1
MM1 pKa = 7.32KK2 pKa = 9.87PIYY5 pKa = 10.24GYY7 pKa = 10.25IFTIVVFVCFIVGFGIADD25 pKa = 3.61NEE27 pKa = 4.16QSMGTSPDD35 pKa = 3.14IPPTTNALSKK45 pKa = 9.6TDD47 pKa = 3.56VVSPSGDD54 pKa = 3.48STGGTDD60 pKa = 3.64SSHH63 pKa = 5.85EE64 pKa = 4.4TTSTTDD70 pKa = 2.8TGSADD75 pKa = 3.41SSSTGQTQTGEE86 pKa = 4.19ITSTTQQSTDD96 pKa = 3.24GLSEE100 pKa = 4.13TGSSSEE106 pKa = 4.31TSSSTTEE113 pKa = 3.73TGSADD118 pKa = 3.15SSTPGQTQTGEE129 pKa = 4.13ITSTTQQSTDD139 pKa = 3.24GLSEE143 pKa = 4.13TGSSSEE149 pKa = 4.31TSSSTTEE156 pKa = 3.79TRR158 pKa = 11.84SADD161 pKa = 3.13ASTTGDD167 pKa = 3.2TTTGQNPSTTEE178 pKa = 3.66HH179 pKa = 5.41TTGDD183 pKa = 3.29VSKK186 pKa = 10.98KK187 pKa = 10.4VGSSLNTASGDD198 pKa = 3.62QTVLNQDD205 pKa = 3.92DD206 pKa = 4.47GTTTSQTVIITPHH219 pKa = 6.38SDD221 pKa = 3.24NDD223 pKa = 3.63EE224 pKa = 4.4KK225 pKa = 11.48SPEE228 pKa = 4.28TVTLGTVTVQSIIDD242 pKa = 3.77LTPSGGTGYY251 pKa = 9.04TYY253 pKa = 11.24DD254 pKa = 4.1STTSSYY260 pKa = 11.34DD261 pKa = 2.84ITGAGNTYY269 pKa = 10.17VLQSGFSTTSTDD281 pKa = 3.51FAIRR285 pKa = 11.84IQASNVILDD294 pKa = 4.05GNYY297 pKa = 7.73QTITGSGGSAGVSVISGTDD316 pKa = 2.99SVTVRR321 pKa = 11.84NFGGINYY328 pKa = 8.18FNNGINSQGDD338 pKa = 3.58STTIFGNTASEE349 pKa = 4.05NNYY352 pKa = 10.49GIVSTGDD359 pKa = 3.05SATISGNTANDD370 pKa = 3.13NGFAGIKK377 pKa = 9.68SDD379 pKa = 3.45GVSAVISGNTANSNDD394 pKa = 3.64EE395 pKa = 4.82GIEE398 pKa = 4.24STGPTATISSNTAEE412 pKa = 4.17NNNNNGINSDD422 pKa = 4.01GEE424 pKa = 4.38SATIRR429 pKa = 11.84SNTVNSNGIAGSDD442 pKa = 3.75CNGIDD447 pKa = 3.85SDD449 pKa = 4.36GNYY452 pKa = 10.66AQISGNIASNNNNNGVDD469 pKa = 4.02SIGVSDD475 pKa = 4.09VIIGNEE481 pKa = 3.77VNNNGKK487 pKa = 10.11SGIYY491 pKa = 9.64SAAPNAFIGTNTADD505 pKa = 3.77GNTQTGIDD513 pKa = 4.03SEE515 pKa = 4.53QTSATISGNTVSNNNYY531 pKa = 9.8GIYY534 pKa = 10.27SAGSDD539 pKa = 3.31AAISSNTANRR549 pKa = 11.84NTIGIVSYY557 pKa = 11.13GDD559 pKa = 3.26SAAINDD565 pKa = 3.56NTANSNNNNGIYY577 pKa = 9.97SDD579 pKa = 4.4GEE581 pKa = 4.3SVTIKK586 pKa = 10.83GNTVAEE592 pKa = 4.29NTNSGLVLGGGSGSVYY608 pKa = 10.71NNKK611 pKa = 9.26FANTNNVGGVSNPSGYY627 pKa = 7.89TWNQAPSPGSNVVGGPNIAGNYY649 pKa = 7.87WSDD652 pKa = 3.4PTGTGWSDD660 pKa = 2.84THH662 pKa = 6.5TPTTTGYY669 pKa = 6.21TTEE672 pKa = 4.31RR673 pKa = 11.84YY674 pKa = 9.17NVGGSMYY681 pKa = 11.16DD682 pKa = 3.25NAPLVKK688 pKa = 9.39TRR690 pKa = 11.84SSPPSPDD697 pKa = 3.77PNPDD701 pKa = 3.46YY702 pKa = 10.96PPSHH706 pKa = 7.47PNQPSDD712 pKa = 3.43PSSRR716 pKa = 11.84SLINIVPNSADD727 pKa = 2.91LSFSVEE733 pKa = 4.06GNQATITLDD742 pKa = 3.41LEE744 pKa = 4.8NIGTVTLSPSARR756 pKa = 11.84IVLVPINDD764 pKa = 3.81PAIPIGVLPASYY776 pKa = 11.02DD777 pKa = 3.42SGKK780 pKa = 10.38YY781 pKa = 10.2LINFGVQIPSSPGTYY796 pKa = 9.08TYY798 pKa = 10.38TFQPEE803 pKa = 4.76MITTDD808 pKa = 3.41PSGNEE813 pKa = 3.64VLMNAGGLVTFIVTVNPDD831 pKa = 2.96GTVNCTLGSS840 pKa = 3.59
MM1 pKa = 7.32KK2 pKa = 9.87PIYY5 pKa = 10.24GYY7 pKa = 10.25IFTIVVFVCFIVGFGIADD25 pKa = 3.61NEE27 pKa = 4.16QSMGTSPDD35 pKa = 3.14IPPTTNALSKK45 pKa = 9.6TDD47 pKa = 3.56VVSPSGDD54 pKa = 3.48STGGTDD60 pKa = 3.64SSHH63 pKa = 5.85EE64 pKa = 4.4TTSTTDD70 pKa = 2.8TGSADD75 pKa = 3.41SSSTGQTQTGEE86 pKa = 4.19ITSTTQQSTDD96 pKa = 3.24GLSEE100 pKa = 4.13TGSSSEE106 pKa = 4.31TSSSTTEE113 pKa = 3.73TGSADD118 pKa = 3.15SSTPGQTQTGEE129 pKa = 4.13ITSTTQQSTDD139 pKa = 3.24GLSEE143 pKa = 4.13TGSSSEE149 pKa = 4.31TSSSTTEE156 pKa = 3.79TRR158 pKa = 11.84SADD161 pKa = 3.13ASTTGDD167 pKa = 3.2TTTGQNPSTTEE178 pKa = 3.66HH179 pKa = 5.41TTGDD183 pKa = 3.29VSKK186 pKa = 10.98KK187 pKa = 10.4VGSSLNTASGDD198 pKa = 3.62QTVLNQDD205 pKa = 3.92DD206 pKa = 4.47GTTTSQTVIITPHH219 pKa = 6.38SDD221 pKa = 3.24NDD223 pKa = 3.63EE224 pKa = 4.4KK225 pKa = 11.48SPEE228 pKa = 4.28TVTLGTVTVQSIIDD242 pKa = 3.77LTPSGGTGYY251 pKa = 9.04TYY253 pKa = 11.24DD254 pKa = 4.1STTSSYY260 pKa = 11.34DD261 pKa = 2.84ITGAGNTYY269 pKa = 10.17VLQSGFSTTSTDD281 pKa = 3.51FAIRR285 pKa = 11.84IQASNVILDD294 pKa = 4.05GNYY297 pKa = 7.73QTITGSGGSAGVSVISGTDD316 pKa = 2.99SVTVRR321 pKa = 11.84NFGGINYY328 pKa = 8.18FNNGINSQGDD338 pKa = 3.58STTIFGNTASEE349 pKa = 4.05NNYY352 pKa = 10.49GIVSTGDD359 pKa = 3.05SATISGNTANDD370 pKa = 3.13NGFAGIKK377 pKa = 9.68SDD379 pKa = 3.45GVSAVISGNTANSNDD394 pKa = 3.64EE395 pKa = 4.82GIEE398 pKa = 4.24STGPTATISSNTAEE412 pKa = 4.17NNNNNGINSDD422 pKa = 4.01GEE424 pKa = 4.38SATIRR429 pKa = 11.84SNTVNSNGIAGSDD442 pKa = 3.75CNGIDD447 pKa = 3.85SDD449 pKa = 4.36GNYY452 pKa = 10.66AQISGNIASNNNNNGVDD469 pKa = 4.02SIGVSDD475 pKa = 4.09VIIGNEE481 pKa = 3.77VNNNGKK487 pKa = 10.11SGIYY491 pKa = 9.64SAAPNAFIGTNTADD505 pKa = 3.77GNTQTGIDD513 pKa = 4.03SEE515 pKa = 4.53QTSATISGNTVSNNNYY531 pKa = 9.8GIYY534 pKa = 10.27SAGSDD539 pKa = 3.31AAISSNTANRR549 pKa = 11.84NTIGIVSYY557 pKa = 11.13GDD559 pKa = 3.26SAAINDD565 pKa = 3.56NTANSNNNNGIYY577 pKa = 9.97SDD579 pKa = 4.4GEE581 pKa = 4.3SVTIKK586 pKa = 10.83GNTVAEE592 pKa = 4.29NTNSGLVLGGGSGSVYY608 pKa = 10.71NNKK611 pKa = 9.26FANTNNVGGVSNPSGYY627 pKa = 7.89TWNQAPSPGSNVVGGPNIAGNYY649 pKa = 7.87WSDD652 pKa = 3.4PTGTGWSDD660 pKa = 2.84THH662 pKa = 6.5TPTTTGYY669 pKa = 6.21TTEE672 pKa = 4.31RR673 pKa = 11.84YY674 pKa = 9.17NVGGSMYY681 pKa = 11.16DD682 pKa = 3.25NAPLVKK688 pKa = 9.39TRR690 pKa = 11.84SSPPSPDD697 pKa = 3.77PNPDD701 pKa = 3.46YY702 pKa = 10.96PPSHH706 pKa = 7.47PNQPSDD712 pKa = 3.43PSSRR716 pKa = 11.84SLINIVPNSADD727 pKa = 2.91LSFSVEE733 pKa = 4.06GNQATITLDD742 pKa = 3.41LEE744 pKa = 4.8NIGTVTLSPSARR756 pKa = 11.84IVLVPINDD764 pKa = 3.81PAIPIGVLPASYY776 pKa = 11.02DD777 pKa = 3.42SGKK780 pKa = 10.38YY781 pKa = 10.2LINFGVQIPSSPGTYY796 pKa = 9.08TYY798 pKa = 10.38TFQPEE803 pKa = 4.76MITTDD808 pKa = 3.41PSGNEE813 pKa = 3.64VLMNAGGLVTFIVTVNPDD831 pKa = 2.96GTVNCTLGSS840 pKa = 3.59
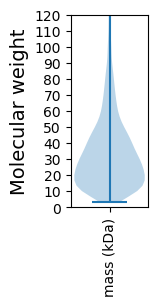
Molecular weight: 85.2 kDa
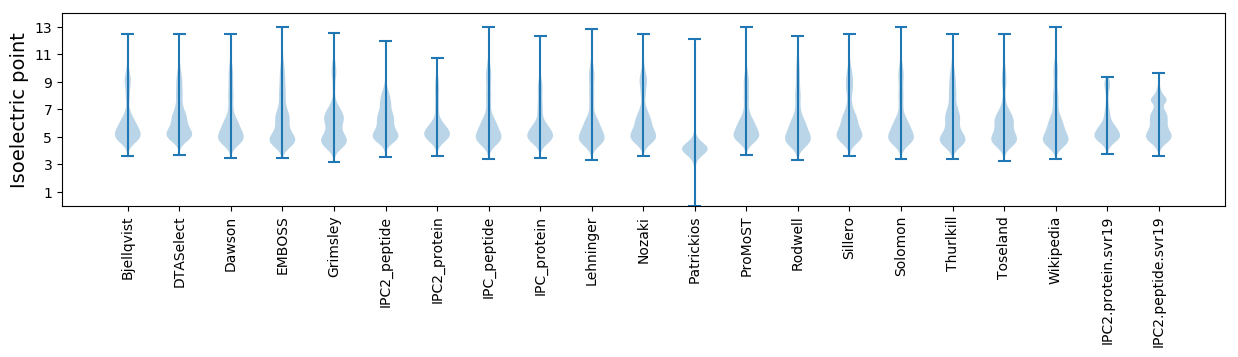
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V2MV55|A0A2V2MV55_9EURY Uncharacterized protein OS=Methanospirillum lacunae OX=668570 GN=DK846_10345 PE=4 SV=1
MM1 pKa = 7.38SKK3 pKa = 10.34VSKK6 pKa = 10.03CRR8 pKa = 11.84KK9 pKa = 8.62IRR11 pKa = 11.84LSKK14 pKa = 9.64KK15 pKa = 6.56TQQNRR20 pKa = 11.84RR21 pKa = 11.84VPQWVMIKK29 pKa = 8.95TKK31 pKa = 10.08RR32 pKa = 11.84KK33 pKa = 10.07VMTHH37 pKa = 6.13PARR40 pKa = 11.84HH41 pKa = 4.6SWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.66VV50 pKa = 3.07
MM1 pKa = 7.38SKK3 pKa = 10.34VSKK6 pKa = 10.03CRR8 pKa = 11.84KK9 pKa = 8.62IRR11 pKa = 11.84LSKK14 pKa = 9.64KK15 pKa = 6.56TQQNRR20 pKa = 11.84RR21 pKa = 11.84VPQWVMIKK29 pKa = 8.95TKK31 pKa = 10.08RR32 pKa = 11.84KK33 pKa = 10.07VMTHH37 pKa = 6.13PARR40 pKa = 11.84HH41 pKa = 4.6SWRR44 pKa = 11.84RR45 pKa = 11.84STLKK49 pKa = 10.66VV50 pKa = 3.07
Molecular weight: 6.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1080887 |
31 |
2928 |
320.1 |
35.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.904 ± 0.047 | 1.332 ± 0.019 |
5.641 ± 0.035 | 6.231 ± 0.046 |
3.954 ± 0.03 | 7.382 ± 0.039 |
2.025 ± 0.02 | 8.638 ± 0.046 |
5.102 ± 0.039 | 9.152 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.53 ± 0.021 | 4.167 ± 0.049 |
4.488 ± 0.03 | 3.276 ± 0.024 |
4.708 ± 0.042 | 7.304 ± 0.047 |
6.006 ± 0.047 | 6.825 ± 0.032 |
1.03 ± 0.015 | 3.302 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
