
Candidatus Terasakiella magnetica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Terasakiellaceae; Terasakiella
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
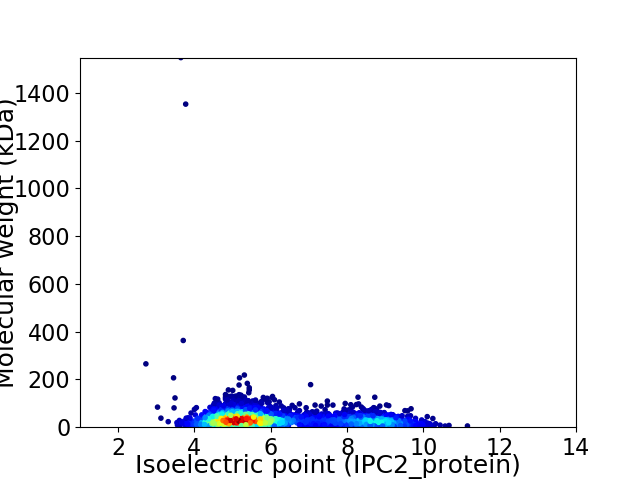
Virtual 2D-PAGE plot for 3572 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
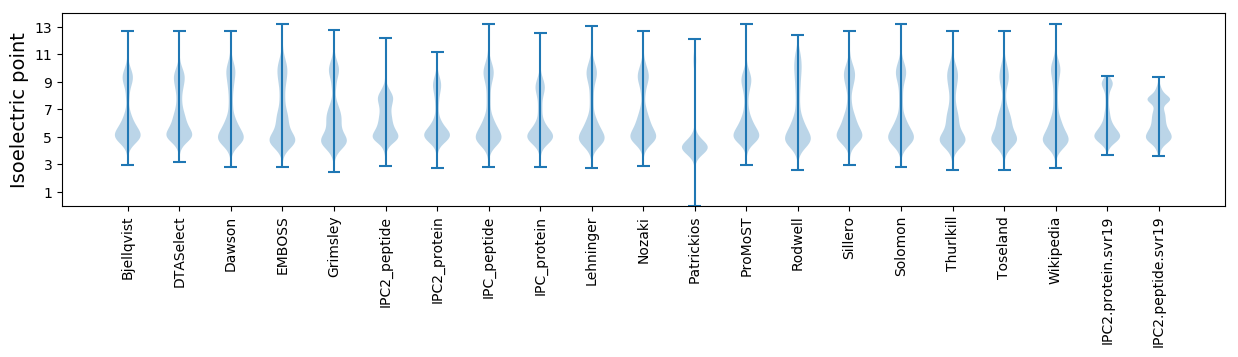
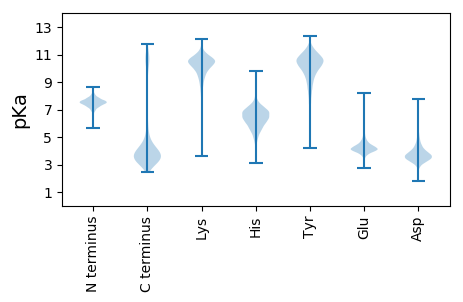
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C3RGI3|A0A1C3RGI3_9PROT Chaperone protein DnaJ OS=Candidatus Terasakiella magnetica OX=1867952 GN=dnaJ PE=3 SV=1
MM1 pKa = 7.66IKK3 pKa = 10.62LLLGTAALSVMSLISVGANAADD25 pKa = 4.66PIKK28 pKa = 10.78LSFSGFMEE36 pKa = 3.77QWVGYY41 pKa = 9.9SDD43 pKa = 4.88NEE45 pKa = 4.24GDD47 pKa = 3.89NGAANYY53 pKa = 10.69SEE55 pKa = 4.79VGTFSDD61 pKa = 4.48VEE63 pKa = 4.29LFMKK67 pKa = 10.83GSTRR71 pKa = 11.84LDD73 pKa = 3.2NGLTVGVNFEE83 pKa = 4.18IEE85 pKa = 4.03RR86 pKa = 11.84SGGADD91 pKa = 3.17GVSDD95 pKa = 3.79EE96 pKa = 5.32SYY98 pKa = 11.71VSVTSDD104 pKa = 2.99ALGTLKK110 pKa = 10.71VGSTMGVSYY119 pKa = 10.91GLSNHH124 pKa = 6.02HH125 pKa = 6.93WDD127 pKa = 3.73VGALMDD133 pKa = 5.89DD134 pKa = 4.17GVHH137 pKa = 6.51QLFAVNALGDD147 pKa = 4.41EE148 pKa = 4.42IDD150 pKa = 3.92TTHH153 pKa = 5.93NTSDD157 pKa = 3.31GHH159 pKa = 6.49KK160 pKa = 10.33VIYY163 pKa = 10.24LSPNLGGVQAGFSYY177 pKa = 11.24GLINEE182 pKa = 4.58TNVGSVDD189 pKa = 3.73TLDD192 pKa = 3.81TNNDD196 pKa = 3.09LQYY199 pKa = 11.27NAGIVYY205 pKa = 10.39SADD208 pKa = 3.48YY209 pKa = 11.0DD210 pKa = 3.97GLSVNVDD217 pKa = 3.17VNYY220 pKa = 10.69EE221 pKa = 4.13LIEE224 pKa = 4.9DD225 pKa = 4.14GGLTGGGVASTTAGSEE241 pKa = 4.07TDD243 pKa = 3.21GTDD246 pKa = 3.04QVKK249 pKa = 10.71NEE251 pKa = 3.98EE252 pKa = 3.89SWRR255 pKa = 11.84AGFTVAQQGLTVSASYY271 pKa = 11.32LEE273 pKa = 4.22TDD275 pKa = 3.23NKK277 pKa = 9.91GTVTGKK283 pKa = 10.49DD284 pKa = 3.07STAWEE289 pKa = 4.12AGVTYY294 pKa = 8.75KK295 pKa = 10.38TGPYY299 pKa = 8.8GFSVGYY305 pKa = 8.76FNRR308 pKa = 11.84ATEE311 pKa = 4.29DD312 pKa = 3.46SGLSTALEE320 pKa = 4.57DD321 pKa = 3.68TTDD324 pKa = 3.39MYY326 pKa = 11.41LVSASYY332 pKa = 11.24NLGPGVTLAGSIVTIEE348 pKa = 4.24TDD350 pKa = 3.55DD351 pKa = 5.19ASDD354 pKa = 3.6SSPNQTDD361 pKa = 3.4EE362 pKa = 4.12GSNWALITGLKK373 pKa = 9.24VHH375 pKa = 6.9FF376 pKa = 4.81
MM1 pKa = 7.66IKK3 pKa = 10.62LLLGTAALSVMSLISVGANAADD25 pKa = 4.66PIKK28 pKa = 10.78LSFSGFMEE36 pKa = 3.77QWVGYY41 pKa = 9.9SDD43 pKa = 4.88NEE45 pKa = 4.24GDD47 pKa = 3.89NGAANYY53 pKa = 10.69SEE55 pKa = 4.79VGTFSDD61 pKa = 4.48VEE63 pKa = 4.29LFMKK67 pKa = 10.83GSTRR71 pKa = 11.84LDD73 pKa = 3.2NGLTVGVNFEE83 pKa = 4.18IEE85 pKa = 4.03RR86 pKa = 11.84SGGADD91 pKa = 3.17GVSDD95 pKa = 3.79EE96 pKa = 5.32SYY98 pKa = 11.71VSVTSDD104 pKa = 2.99ALGTLKK110 pKa = 10.71VGSTMGVSYY119 pKa = 10.91GLSNHH124 pKa = 6.02HH125 pKa = 6.93WDD127 pKa = 3.73VGALMDD133 pKa = 5.89DD134 pKa = 4.17GVHH137 pKa = 6.51QLFAVNALGDD147 pKa = 4.41EE148 pKa = 4.42IDD150 pKa = 3.92TTHH153 pKa = 5.93NTSDD157 pKa = 3.31GHH159 pKa = 6.49KK160 pKa = 10.33VIYY163 pKa = 10.24LSPNLGGVQAGFSYY177 pKa = 11.24GLINEE182 pKa = 4.58TNVGSVDD189 pKa = 3.73TLDD192 pKa = 3.81TNNDD196 pKa = 3.09LQYY199 pKa = 11.27NAGIVYY205 pKa = 10.39SADD208 pKa = 3.48YY209 pKa = 11.0DD210 pKa = 3.97GLSVNVDD217 pKa = 3.17VNYY220 pKa = 10.69EE221 pKa = 4.13LIEE224 pKa = 4.9DD225 pKa = 4.14GGLTGGGVASTTAGSEE241 pKa = 4.07TDD243 pKa = 3.21GTDD246 pKa = 3.04QVKK249 pKa = 10.71NEE251 pKa = 3.98EE252 pKa = 3.89SWRR255 pKa = 11.84AGFTVAQQGLTVSASYY271 pKa = 11.32LEE273 pKa = 4.22TDD275 pKa = 3.23NKK277 pKa = 9.91GTVTGKK283 pKa = 10.49DD284 pKa = 3.07STAWEE289 pKa = 4.12AGVTYY294 pKa = 8.75KK295 pKa = 10.38TGPYY299 pKa = 8.8GFSVGYY305 pKa = 8.76FNRR308 pKa = 11.84ATEE311 pKa = 4.29DD312 pKa = 3.46SGLSTALEE320 pKa = 4.57DD321 pKa = 3.68TTDD324 pKa = 3.39MYY326 pKa = 11.41LVSASYY332 pKa = 11.24NLGPGVTLAGSIVTIEE348 pKa = 4.24TDD350 pKa = 3.55DD351 pKa = 5.19ASDD354 pKa = 3.6SSPNQTDD361 pKa = 3.4EE362 pKa = 4.12GSNWALITGLKK373 pKa = 9.24VHH375 pKa = 6.9FF376 pKa = 4.81
Molecular weight: 39.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C3RHC1|A0A1C3RHC1_9PROT Putative D-alanine--D-alanine ligase domain protein OS=Candidatus Terasakiella magnetica OX=1867952 GN=MTBPR1_260007 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVAGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.97GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVAGRR28 pKa = 11.84RR29 pKa = 11.84VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.97GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1122897 |
21 |
14707 |
314.4 |
34.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.976 ± 0.062 | 1.063 ± 0.024 |
6.14 ± 0.082 | 6.955 ± 0.053 |
4.053 ± 0.029 | 7.384 ± 0.058 |
2.238 ± 0.027 | 6.19 ± 0.036 |
5.897 ± 0.069 | 9.66 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.84 ± 0.037 | 3.863 ± 0.048 |
3.999 ± 0.048 | 3.693 ± 0.027 |
4.893 ± 0.072 | 6.088 ± 0.056 |
5.314 ± 0.086 | 6.92 ± 0.034 |
1.144 ± 0.016 | 2.691 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
