
unidentified eubacterium SCB49
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; environmental samples
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
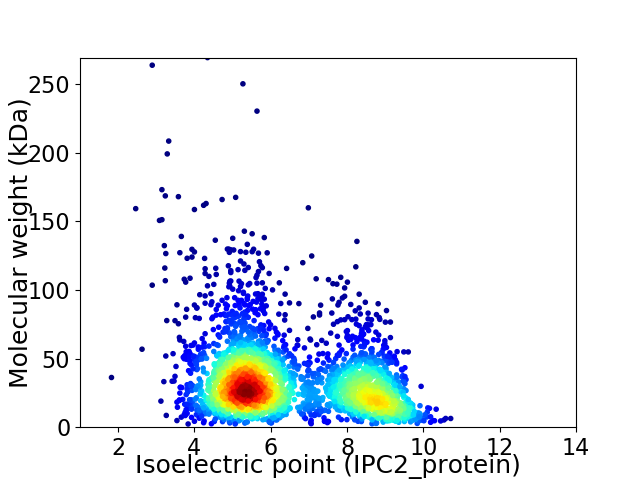
Virtual 2D-PAGE plot for 2926 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6ETP2|A6ETP2_9BACT 3-dehydroquinate dehydratase OS=unidentified eubacterium SCB49 OX=50743 GN=aroQ PE=3 SV=1
MM1 pKa = 7.37KK2 pKa = 10.25KK3 pKa = 9.96IYY5 pKa = 10.17LLLGLCPILSVQSQNFEE22 pKa = 4.09EE23 pKa = 5.17VEE25 pKa = 4.03TDD27 pKa = 3.15MKK29 pKa = 10.92DD30 pKa = 3.44YY31 pKa = 10.79YY32 pKa = 11.11YY33 pKa = 11.14SSSNVADD40 pKa = 3.92FDD42 pKa = 4.47GDD44 pKa = 3.94GFVDD48 pKa = 3.51IVFNGAIDD56 pKa = 3.65SDD58 pKa = 4.31NNGEE62 pKa = 3.92VDD64 pKa = 3.42TTFNEE69 pKa = 4.71VYY71 pKa = 10.59KK72 pKa = 11.04NDD74 pKa = 4.01DD75 pKa = 3.23ATFTLFDD82 pKa = 4.74DD83 pKa = 4.45LGLYY87 pKa = 7.63ATHH90 pKa = 7.11LGDD93 pKa = 5.26IKK95 pKa = 11.08FLDD98 pKa = 3.98FDD100 pKa = 4.29NDD102 pKa = 3.49GLLDD106 pKa = 3.67IVSTGLSYY114 pKa = 11.72NDD116 pKa = 2.86ITDD119 pKa = 3.59YY120 pKa = 10.35QQYY123 pKa = 10.27RR124 pKa = 11.84LKK126 pKa = 10.2NTGTSLEE133 pKa = 4.19LEE135 pKa = 4.31EE136 pKa = 5.26NISGKK141 pKa = 9.89IFGSLEE147 pKa = 4.01VFDD150 pKa = 5.28FNHH153 pKa = 7.09DD154 pKa = 3.75GFVDD158 pKa = 3.73YY159 pKa = 10.89ALNGIQYY166 pKa = 9.77IEE168 pKa = 4.27EE169 pKa = 4.24DD170 pKa = 3.47QAFSFDD176 pKa = 4.26LDD178 pKa = 4.47FYY180 pKa = 11.33QNNGDD185 pKa = 4.03GFDD188 pKa = 3.76VTPAWLSGTQSGAFKK203 pKa = 11.06VLDD206 pKa = 4.22LNNDD210 pKa = 3.23NLLDD214 pKa = 3.88LVISGFNIDD223 pKa = 3.93YY224 pKa = 10.76EE225 pKa = 4.75PIFKK229 pKa = 10.61VYY231 pKa = 10.83LNNNGVLEE239 pKa = 4.43FSQDD243 pKa = 3.42LAPISDD249 pKa = 3.9GEE251 pKa = 4.05IAYY254 pKa = 9.23TDD256 pKa = 4.19FNADD260 pKa = 3.13GFLDD264 pKa = 3.93LAVVGVDD271 pKa = 3.82LDD273 pKa = 4.07YY274 pKa = 11.9NEE276 pKa = 4.28YY277 pKa = 10.68LAVLINDD284 pKa = 3.55GTGNFTTNVIDD295 pKa = 3.74NEE297 pKa = 4.53GVSGSSIDD305 pKa = 4.88VGDD308 pKa = 4.45LNNDD312 pKa = 3.22GYY314 pKa = 11.7YY315 pKa = 10.95DD316 pKa = 4.52FIIIGNDD323 pKa = 2.68ADD325 pKa = 3.86YY326 pKa = 11.29NGWVKK331 pKa = 10.49VFLYY335 pKa = 10.91DD336 pKa = 3.91NNTEE340 pKa = 4.32SFTKK344 pKa = 10.67SADD347 pKa = 3.18TGLYY351 pKa = 10.41NLGSNGDD358 pKa = 3.6INLFDD363 pKa = 4.37YY364 pKa = 11.59NNDD367 pKa = 2.97NHH369 pKa = 7.86LDD371 pKa = 3.56VMMSGFDD378 pKa = 3.28WADD381 pKa = 2.84SDD383 pKa = 5.06FPSLTKK389 pKa = 10.86LFTNLSTEE397 pKa = 4.42EE398 pKa = 4.06NQKK401 pKa = 10.03PLPPTEE407 pKa = 5.28LDD409 pKa = 3.4LTQDD413 pKa = 3.48DD414 pKa = 4.07NKK416 pKa = 10.25YY417 pKa = 7.91TFTWSGASDD426 pKa = 4.02DD427 pKa = 4.03KK428 pKa = 11.52TPTNALQYY436 pKa = 8.68EE437 pKa = 4.6IKK439 pKa = 10.89VGTTSGAQDD448 pKa = 3.24VAKK451 pKa = 10.76YY452 pKa = 10.25IVTTPSWFLEE462 pKa = 4.21LEE464 pKa = 4.11NMPEE468 pKa = 3.67NLYY471 pKa = 10.36WSVKK475 pKa = 10.62SIDD478 pKa = 3.35ASKK481 pKa = 10.57ILSNSSDD488 pKa = 3.6EE489 pKa = 4.29QQLSVSDD496 pKa = 4.75FSLLNNIKK504 pKa = 10.23VYY506 pKa = 10.59PNPASGKK513 pKa = 8.56VFVSAEE519 pKa = 3.74GLIAVEE525 pKa = 4.76MYY527 pKa = 10.37DD528 pKa = 3.3IQGRR532 pKa = 11.84KK533 pKa = 9.45INVKK537 pKa = 10.25LNSDD541 pKa = 4.42FSLDD545 pKa = 3.31ISGLSSGVYY554 pKa = 7.92VLKK557 pKa = 10.79MNVDD561 pKa = 3.55GVLVSRR567 pKa = 11.84KK568 pKa = 10.06LSVNN572 pKa = 3.27
MM1 pKa = 7.37KK2 pKa = 10.25KK3 pKa = 9.96IYY5 pKa = 10.17LLLGLCPILSVQSQNFEE22 pKa = 4.09EE23 pKa = 5.17VEE25 pKa = 4.03TDD27 pKa = 3.15MKK29 pKa = 10.92DD30 pKa = 3.44YY31 pKa = 10.79YY32 pKa = 11.11YY33 pKa = 11.14SSSNVADD40 pKa = 3.92FDD42 pKa = 4.47GDD44 pKa = 3.94GFVDD48 pKa = 3.51IVFNGAIDD56 pKa = 3.65SDD58 pKa = 4.31NNGEE62 pKa = 3.92VDD64 pKa = 3.42TTFNEE69 pKa = 4.71VYY71 pKa = 10.59KK72 pKa = 11.04NDD74 pKa = 4.01DD75 pKa = 3.23ATFTLFDD82 pKa = 4.74DD83 pKa = 4.45LGLYY87 pKa = 7.63ATHH90 pKa = 7.11LGDD93 pKa = 5.26IKK95 pKa = 11.08FLDD98 pKa = 3.98FDD100 pKa = 4.29NDD102 pKa = 3.49GLLDD106 pKa = 3.67IVSTGLSYY114 pKa = 11.72NDD116 pKa = 2.86ITDD119 pKa = 3.59YY120 pKa = 10.35QQYY123 pKa = 10.27RR124 pKa = 11.84LKK126 pKa = 10.2NTGTSLEE133 pKa = 4.19LEE135 pKa = 4.31EE136 pKa = 5.26NISGKK141 pKa = 9.89IFGSLEE147 pKa = 4.01VFDD150 pKa = 5.28FNHH153 pKa = 7.09DD154 pKa = 3.75GFVDD158 pKa = 3.73YY159 pKa = 10.89ALNGIQYY166 pKa = 9.77IEE168 pKa = 4.27EE169 pKa = 4.24DD170 pKa = 3.47QAFSFDD176 pKa = 4.26LDD178 pKa = 4.47FYY180 pKa = 11.33QNNGDD185 pKa = 4.03GFDD188 pKa = 3.76VTPAWLSGTQSGAFKK203 pKa = 11.06VLDD206 pKa = 4.22LNNDD210 pKa = 3.23NLLDD214 pKa = 3.88LVISGFNIDD223 pKa = 3.93YY224 pKa = 10.76EE225 pKa = 4.75PIFKK229 pKa = 10.61VYY231 pKa = 10.83LNNNGVLEE239 pKa = 4.43FSQDD243 pKa = 3.42LAPISDD249 pKa = 3.9GEE251 pKa = 4.05IAYY254 pKa = 9.23TDD256 pKa = 4.19FNADD260 pKa = 3.13GFLDD264 pKa = 3.93LAVVGVDD271 pKa = 3.82LDD273 pKa = 4.07YY274 pKa = 11.9NEE276 pKa = 4.28YY277 pKa = 10.68LAVLINDD284 pKa = 3.55GTGNFTTNVIDD295 pKa = 3.74NEE297 pKa = 4.53GVSGSSIDD305 pKa = 4.88VGDD308 pKa = 4.45LNNDD312 pKa = 3.22GYY314 pKa = 11.7YY315 pKa = 10.95DD316 pKa = 4.52FIIIGNDD323 pKa = 2.68ADD325 pKa = 3.86YY326 pKa = 11.29NGWVKK331 pKa = 10.49VFLYY335 pKa = 10.91DD336 pKa = 3.91NNTEE340 pKa = 4.32SFTKK344 pKa = 10.67SADD347 pKa = 3.18TGLYY351 pKa = 10.41NLGSNGDD358 pKa = 3.6INLFDD363 pKa = 4.37YY364 pKa = 11.59NNDD367 pKa = 2.97NHH369 pKa = 7.86LDD371 pKa = 3.56VMMSGFDD378 pKa = 3.28WADD381 pKa = 2.84SDD383 pKa = 5.06FPSLTKK389 pKa = 10.86LFTNLSTEE397 pKa = 4.42EE398 pKa = 4.06NQKK401 pKa = 10.03PLPPTEE407 pKa = 5.28LDD409 pKa = 3.4LTQDD413 pKa = 3.48DD414 pKa = 4.07NKK416 pKa = 10.25YY417 pKa = 7.91TFTWSGASDD426 pKa = 4.02DD427 pKa = 4.03KK428 pKa = 11.52TPTNALQYY436 pKa = 8.68EE437 pKa = 4.6IKK439 pKa = 10.89VGTTSGAQDD448 pKa = 3.24VAKK451 pKa = 10.76YY452 pKa = 10.25IVTTPSWFLEE462 pKa = 4.21LEE464 pKa = 4.11NMPEE468 pKa = 3.67NLYY471 pKa = 10.36WSVKK475 pKa = 10.62SIDD478 pKa = 3.35ASKK481 pKa = 10.57ILSNSSDD488 pKa = 3.6EE489 pKa = 4.29QQLSVSDD496 pKa = 4.75FSLLNNIKK504 pKa = 10.23VYY506 pKa = 10.59PNPASGKK513 pKa = 8.56VFVSAEE519 pKa = 3.74GLIAVEE525 pKa = 4.76MYY527 pKa = 10.37DD528 pKa = 3.3IQGRR532 pKa = 11.84KK533 pKa = 9.45INVKK537 pKa = 10.25LNSDD541 pKa = 4.42FSLDD545 pKa = 3.31ISGLSSGVYY554 pKa = 7.92VLKK557 pKa = 10.79MNVDD561 pKa = 3.55GVLVSRR567 pKa = 11.84KK568 pKa = 10.06LSVNN572 pKa = 3.27
Molecular weight: 63.53 kDa
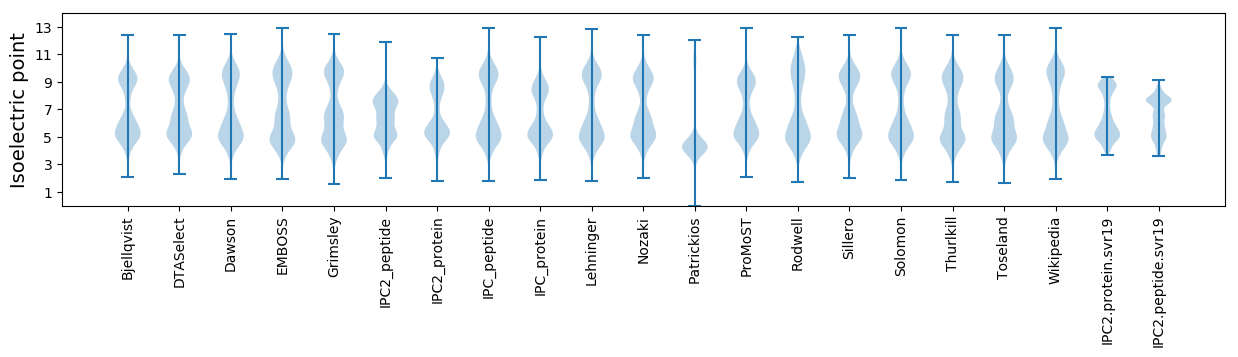
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6ESE5|A6ESE5_9BACT N-acetyltransferase domain-containing protein OS=unidentified eubacterium SCB49 OX=50743 GN=SCB49_00205 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.17RR22 pKa = 11.84MASVSGRR29 pKa = 11.84KK30 pKa = 9.02VIKK33 pKa = 10.15ARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.71GRR40 pKa = 11.84KK41 pKa = 8.56KK42 pKa = 10.24ISVSSEE48 pKa = 3.49FRR50 pKa = 11.84HH51 pKa = 6.21KK52 pKa = 10.57KK53 pKa = 9.45
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.17RR22 pKa = 11.84MASVSGRR29 pKa = 11.84KK30 pKa = 9.02VIKK33 pKa = 10.15ARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.71GRR40 pKa = 11.84KK41 pKa = 8.56KK42 pKa = 10.24ISVSSEE48 pKa = 3.49FRR50 pKa = 11.84HH51 pKa = 6.21KK52 pKa = 10.57KK53 pKa = 9.45
Molecular weight: 6.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
950377 |
21 |
2545 |
324.8 |
36.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.794 ± 0.048 | 0.765 ± 0.018 |
5.85 ± 0.054 | 6.662 ± 0.043 |
5.125 ± 0.034 | 6.438 ± 0.049 |
1.698 ± 0.019 | 7.854 ± 0.042 |
7.58 ± 0.064 | 9.173 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.169 ± 0.021 | 5.981 ± 0.05 |
3.325 ± 0.028 | 3.323 ± 0.023 |
3.269 ± 0.034 | 6.372 ± 0.034 |
6.254 ± 0.046 | 6.427 ± 0.036 |
0.973 ± 0.016 | 3.967 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
