
Marek s disease virus serotype 2 MDV2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Mardivirus; Gallid alphaherpesvirus 3
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
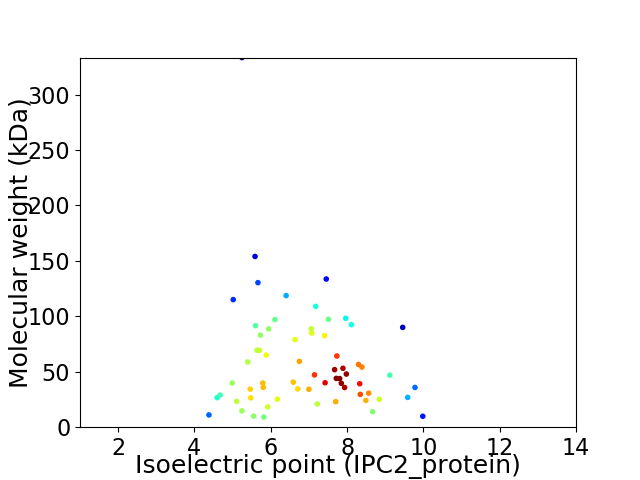
Virtual 2D-PAGE plot for 65 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1P7U0A2|A0A1P7U0A2_9ALPH Capsid vertex component 1 OS=Marek's disease virus serotype 2 MDV2 OX=36353 GN=ORF 22 PE=3 SV=1
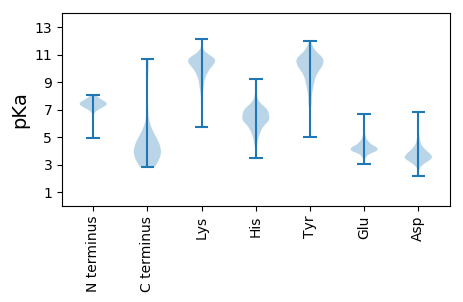
MM1 pKa = 6.81PWYY4 pKa = 10.2RR5 pKa = 11.84RR6 pKa = 11.84GYY8 pKa = 8.48HH9 pKa = 4.6TTKK12 pKa = 9.62PTDD15 pKa = 3.42CDD17 pKa = 3.21ILLDD21 pKa = 4.21YY22 pKa = 11.04IGDD25 pKa = 3.93EE26 pKa = 3.99QLEE29 pKa = 4.55TNMQDD34 pKa = 2.99TEE36 pKa = 4.99SIVTRR41 pKa = 11.84VSDD44 pKa = 6.25DD45 pKa = 3.42IDD47 pKa = 5.28CEE49 pKa = 4.14DD50 pKa = 3.59AAEE53 pKa = 4.04YY54 pKa = 8.89ATMSSYY60 pKa = 11.33LSGSIFTISKK70 pKa = 9.63SCDD73 pKa = 3.23MNPKK77 pKa = 7.36FTRR80 pKa = 11.84YY81 pKa = 8.98VVLSWLSAFVLRR93 pKa = 11.84PSCCIIFLVYY103 pKa = 11.09AMDD106 pKa = 3.56GHH108 pKa = 6.9DD109 pKa = 3.95NRR111 pKa = 11.84FLILGATITAVFYY124 pKa = 8.44GTLLLEE130 pKa = 4.33TYY132 pKa = 10.92YY133 pKa = 10.11MYY135 pKa = 11.14RR136 pKa = 11.84NIKK139 pKa = 9.27NDD141 pKa = 3.56VMPMDD146 pKa = 4.13NCQQIFVGILSTLGAVIFGVFSYY169 pKa = 11.09RR170 pKa = 11.84MVFQDD175 pKa = 3.42PRR177 pKa = 11.84FMEE180 pKa = 4.29KK181 pKa = 10.24VLQLEE186 pKa = 4.69EE187 pKa = 3.97NDD189 pKa = 3.75KK190 pKa = 10.87TDD192 pKa = 3.17GAVVYY197 pKa = 10.05LLMGATITYY206 pKa = 7.92ATVAVSDD213 pKa = 3.92ALGFLLPRR221 pKa = 11.84LWTRR225 pKa = 11.84ALIKK229 pKa = 10.01TCVPFF234 pKa = 5.21
MM1 pKa = 6.81PWYY4 pKa = 10.2RR5 pKa = 11.84RR6 pKa = 11.84GYY8 pKa = 8.48HH9 pKa = 4.6TTKK12 pKa = 9.62PTDD15 pKa = 3.42CDD17 pKa = 3.21ILLDD21 pKa = 4.21YY22 pKa = 11.04IGDD25 pKa = 3.93EE26 pKa = 3.99QLEE29 pKa = 4.55TNMQDD34 pKa = 2.99TEE36 pKa = 4.99SIVTRR41 pKa = 11.84VSDD44 pKa = 6.25DD45 pKa = 3.42IDD47 pKa = 5.28CEE49 pKa = 4.14DD50 pKa = 3.59AAEE53 pKa = 4.04YY54 pKa = 8.89ATMSSYY60 pKa = 11.33LSGSIFTISKK70 pKa = 9.63SCDD73 pKa = 3.23MNPKK77 pKa = 7.36FTRR80 pKa = 11.84YY81 pKa = 8.98VVLSWLSAFVLRR93 pKa = 11.84PSCCIIFLVYY103 pKa = 11.09AMDD106 pKa = 3.56GHH108 pKa = 6.9DD109 pKa = 3.95NRR111 pKa = 11.84FLILGATITAVFYY124 pKa = 8.44GTLLLEE130 pKa = 4.33TYY132 pKa = 10.92YY133 pKa = 10.11MYY135 pKa = 11.14RR136 pKa = 11.84NIKK139 pKa = 9.27NDD141 pKa = 3.56VMPMDD146 pKa = 4.13NCQQIFVGILSTLGAVIFGVFSYY169 pKa = 11.09RR170 pKa = 11.84MVFQDD175 pKa = 3.42PRR177 pKa = 11.84FMEE180 pKa = 4.29KK181 pKa = 10.24VLQLEE186 pKa = 4.69EE187 pKa = 3.97NDD189 pKa = 3.75KK190 pKa = 10.87TDD192 pKa = 3.17GAVVYY197 pKa = 10.05LLMGATITYY206 pKa = 7.92ATVAVSDD213 pKa = 3.92ALGFLLPRR221 pKa = 11.84LWTRR225 pKa = 11.84ALIKK229 pKa = 10.01TCVPFF234 pKa = 5.21
Molecular weight: 26.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O89248|O89248_9ALPH Membrane protein OS=Marek's disease virus serotype 2 MDV2 OX=36353 GN=UL49.5 PE=3 SV=1
MM1 pKa = 7.7GDD3 pKa = 3.1SDD5 pKa = 3.61RR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.0SSRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84STMRR17 pKa = 11.84TSPDD21 pKa = 2.86NSAHH25 pKa = 6.24ISSTRR30 pKa = 11.84ARR32 pKa = 11.84RR33 pKa = 11.84DD34 pKa = 3.04SSKK37 pKa = 11.15NEE39 pKa = 3.82SPDD42 pKa = 3.78RR43 pKa = 11.84ISPPSHH49 pKa = 5.78SLQRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84SVKK58 pKa = 9.89IEE60 pKa = 4.0RR61 pKa = 11.84KK62 pKa = 9.71DD63 pKa = 3.53SSSEE67 pKa = 4.02TQRR70 pKa = 11.84GEE72 pKa = 4.1SLSSKK77 pKa = 9.87VRR79 pKa = 11.84AKK81 pKa = 10.36PGARR85 pKa = 11.84AIEE88 pKa = 4.05KK89 pKa = 10.6GKK91 pKa = 10.1FAFSTTPASATSTWRR106 pKa = 11.84SNTLVYY112 pKa = 10.61NEE114 pKa = 5.31RR115 pKa = 11.84IFCGAVAAVAQYY127 pKa = 10.11HH128 pKa = 7.1AYY130 pKa = 10.29RR131 pKa = 11.84GALSLWRR138 pKa = 11.84RR139 pKa = 11.84NAPRR143 pKa = 11.84TNAEE147 pKa = 3.97LEE149 pKa = 4.22EE150 pKa = 4.1FLARR154 pKa = 11.84AIIKK158 pKa = 8.5ITIQEE163 pKa = 4.25GANLLDD169 pKa = 3.83EE170 pKa = 5.86AEE172 pKa = 4.13ACTRR176 pKa = 11.84KK177 pKa = 10.16LSEE180 pKa = 4.29EE181 pKa = 4.33SGLSPDD187 pKa = 3.2MGNPKK192 pKa = 9.99SRR194 pKa = 11.84SQYY197 pKa = 9.87GKK199 pKa = 10.05RR200 pKa = 11.84DD201 pKa = 3.55GDD203 pKa = 3.35EE204 pKa = 3.97STPVDD209 pKa = 3.36KK210 pKa = 10.49RR211 pKa = 11.84DD212 pKa = 3.64RR213 pKa = 11.84RR214 pKa = 11.84SKK216 pKa = 9.51TPGRR220 pKa = 11.84APTTSRR226 pKa = 11.84RR227 pKa = 11.84HH228 pKa = 4.88YY229 pKa = 10.07SSSRR233 pKa = 11.84GNYY236 pKa = 9.31SSEE239 pKa = 4.21SEE241 pKa = 4.1
MM1 pKa = 7.7GDD3 pKa = 3.1SDD5 pKa = 3.61RR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.0SSRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84STMRR17 pKa = 11.84TSPDD21 pKa = 2.86NSAHH25 pKa = 6.24ISSTRR30 pKa = 11.84ARR32 pKa = 11.84RR33 pKa = 11.84DD34 pKa = 3.04SSKK37 pKa = 11.15NEE39 pKa = 3.82SPDD42 pKa = 3.78RR43 pKa = 11.84ISPPSHH49 pKa = 5.78SLQRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84SVKK58 pKa = 9.89IEE60 pKa = 4.0RR61 pKa = 11.84KK62 pKa = 9.71DD63 pKa = 3.53SSSEE67 pKa = 4.02TQRR70 pKa = 11.84GEE72 pKa = 4.1SLSSKK77 pKa = 9.87VRR79 pKa = 11.84AKK81 pKa = 10.36PGARR85 pKa = 11.84AIEE88 pKa = 4.05KK89 pKa = 10.6GKK91 pKa = 10.1FAFSTTPASATSTWRR106 pKa = 11.84SNTLVYY112 pKa = 10.61NEE114 pKa = 5.31RR115 pKa = 11.84IFCGAVAAVAQYY127 pKa = 10.11HH128 pKa = 7.1AYY130 pKa = 10.29RR131 pKa = 11.84GALSLWRR138 pKa = 11.84RR139 pKa = 11.84NAPRR143 pKa = 11.84TNAEE147 pKa = 3.97LEE149 pKa = 4.22EE150 pKa = 4.1FLARR154 pKa = 11.84AIIKK158 pKa = 8.5ITIQEE163 pKa = 4.25GANLLDD169 pKa = 3.83EE170 pKa = 5.86AEE172 pKa = 4.13ACTRR176 pKa = 11.84KK177 pKa = 10.16LSEE180 pKa = 4.29EE181 pKa = 4.33SGLSPDD187 pKa = 3.2MGNPKK192 pKa = 9.99SRR194 pKa = 11.84SQYY197 pKa = 9.87GKK199 pKa = 10.05RR200 pKa = 11.84DD201 pKa = 3.55GDD203 pKa = 3.35EE204 pKa = 3.97STPVDD209 pKa = 3.36KK210 pKa = 10.49RR211 pKa = 11.84DD212 pKa = 3.64RR213 pKa = 11.84RR214 pKa = 11.84SKK216 pKa = 9.51TPGRR220 pKa = 11.84APTTSRR226 pKa = 11.84RR227 pKa = 11.84HH228 pKa = 4.88YY229 pKa = 10.07SSSRR233 pKa = 11.84GNYY236 pKa = 9.31SSEE239 pKa = 4.21SEE241 pKa = 4.1
Molecular weight: 26.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
34710 |
81 |
3064 |
534.0 |
59.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.199 ± 0.28 | 2.371 ± 0.163 |
5.725 ± 0.198 | 5.246 ± 0.139 |
3.881 ± 0.178 | 5.938 ± 0.143 |
2.285 ± 0.095 | 5.454 ± 0.155 |
3.786 ± 0.158 | 9.476 ± 0.147 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.1 | 3.584 ± 0.111 |
5.056 ± 0.204 | 2.633 ± 0.11 |
7.312 ± 0.264 | 8.459 ± 0.258 |
6.384 ± 0.156 | 6.502 ± 0.152 |
1.066 ± 0.067 | 3.192 ± 0.12 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
