
Peruvian horse sickness virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
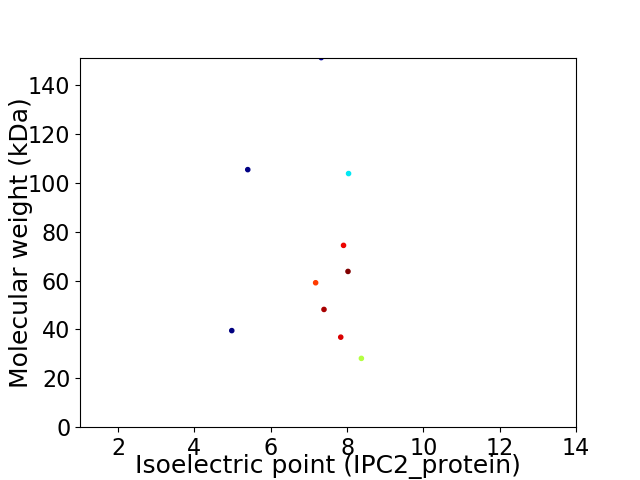
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2Q1D9|Q2Q1D9_9REOV VP6 OS=Peruvian horse sickness virus OX=356862 PE=4 SV=1
MM1 pKa = 7.27GGIYY5 pKa = 10.42ARR7 pKa = 11.84TFCYY11 pKa = 10.48LEE13 pKa = 4.3CLASNRR19 pKa = 11.84DD20 pKa = 3.11PRR22 pKa = 11.84ARR24 pKa = 11.84RR25 pKa = 11.84TFLPSDD31 pKa = 4.27GFPLFVTRR39 pKa = 11.84FNATTNRR46 pKa = 11.84PITQTPSTPEE56 pKa = 3.37EE57 pKa = 3.97HH58 pKa = 6.85RR59 pKa = 11.84ACFYY63 pKa = 11.24AALDD67 pKa = 4.08LLIGATGINFNYY79 pKa = 10.16RR80 pKa = 11.84LPEE83 pKa = 3.87YY84 pKa = 10.35SPNVQVLSILARR96 pKa = 11.84DD97 pKa = 3.85DD98 pKa = 4.38LPYY101 pKa = 10.35TSEE104 pKa = 3.84AFIRR108 pKa = 11.84ATRR111 pKa = 11.84ITNEE115 pKa = 3.68TAAGQSTRR123 pKa = 11.84RR124 pKa = 11.84FNLPWPDD131 pKa = 3.21VNFIYY136 pKa = 10.35PPATPYY142 pKa = 9.65RR143 pKa = 11.84TVDD146 pKa = 3.66DD147 pKa = 4.16QPQPMVIGPQDD158 pKa = 3.41MEE160 pKa = 3.64ITLNVNEE167 pKa = 4.9DD168 pKa = 3.62VEE170 pKa = 4.37ITDD173 pKa = 3.27IVMPNYY179 pKa = 9.54PDD181 pKa = 4.02VINTCFVWYY190 pKa = 8.75TLSHH194 pKa = 5.47YY195 pKa = 10.41RR196 pKa = 11.84GAAGALVEE204 pKa = 5.53GSQQCSVTAGDD215 pKa = 4.22EE216 pKa = 4.15EE217 pKa = 5.09AEE219 pKa = 4.28SGVEE223 pKa = 3.96IIVPAGTAIRR233 pKa = 11.84VSNTSAVYY241 pKa = 10.68AGMIHH246 pKa = 7.08FNIRR250 pKa = 11.84WFTRR254 pKa = 11.84AQPSEE259 pKa = 4.15DD260 pKa = 4.36LYY262 pKa = 11.3DD263 pKa = 3.54TMRR266 pKa = 11.84VDD268 pKa = 4.88FAASYY273 pKa = 10.67SFHH276 pKa = 7.36SEE278 pKa = 2.61QWYY281 pKa = 10.02NLRR284 pKa = 11.84SYY286 pKa = 11.18LLRR289 pKa = 11.84LIGLPPHH296 pKa = 7.12IPPNDD301 pKa = 4.74PIRR304 pKa = 11.84VPNRR308 pKa = 11.84VLMLALISRR317 pKa = 11.84LQDD320 pKa = 2.89VYY322 pKa = 11.13VAHH325 pKa = 6.58SPIDD329 pKa = 3.21IVAQQEE335 pKa = 4.61GNGQAGLPDD344 pKa = 3.83AALEE348 pKa = 3.95ALRR351 pKa = 11.84RR352 pKa = 11.84VV353 pKa = 3.65
MM1 pKa = 7.27GGIYY5 pKa = 10.42ARR7 pKa = 11.84TFCYY11 pKa = 10.48LEE13 pKa = 4.3CLASNRR19 pKa = 11.84DD20 pKa = 3.11PRR22 pKa = 11.84ARR24 pKa = 11.84RR25 pKa = 11.84TFLPSDD31 pKa = 4.27GFPLFVTRR39 pKa = 11.84FNATTNRR46 pKa = 11.84PITQTPSTPEE56 pKa = 3.37EE57 pKa = 3.97HH58 pKa = 6.85RR59 pKa = 11.84ACFYY63 pKa = 11.24AALDD67 pKa = 4.08LLIGATGINFNYY79 pKa = 10.16RR80 pKa = 11.84LPEE83 pKa = 3.87YY84 pKa = 10.35SPNVQVLSILARR96 pKa = 11.84DD97 pKa = 3.85DD98 pKa = 4.38LPYY101 pKa = 10.35TSEE104 pKa = 3.84AFIRR108 pKa = 11.84ATRR111 pKa = 11.84ITNEE115 pKa = 3.68TAAGQSTRR123 pKa = 11.84RR124 pKa = 11.84FNLPWPDD131 pKa = 3.21VNFIYY136 pKa = 10.35PPATPYY142 pKa = 9.65RR143 pKa = 11.84TVDD146 pKa = 3.66DD147 pKa = 4.16QPQPMVIGPQDD158 pKa = 3.41MEE160 pKa = 3.64ITLNVNEE167 pKa = 4.9DD168 pKa = 3.62VEE170 pKa = 4.37ITDD173 pKa = 3.27IVMPNYY179 pKa = 9.54PDD181 pKa = 4.02VINTCFVWYY190 pKa = 8.75TLSHH194 pKa = 5.47YY195 pKa = 10.41RR196 pKa = 11.84GAAGALVEE204 pKa = 5.53GSQQCSVTAGDD215 pKa = 4.22EE216 pKa = 4.15EE217 pKa = 5.09AEE219 pKa = 4.28SGVEE223 pKa = 3.96IIVPAGTAIRR233 pKa = 11.84VSNTSAVYY241 pKa = 10.68AGMIHH246 pKa = 7.08FNIRR250 pKa = 11.84WFTRR254 pKa = 11.84AQPSEE259 pKa = 4.15DD260 pKa = 4.36LYY262 pKa = 11.3DD263 pKa = 3.54TMRR266 pKa = 11.84VDD268 pKa = 4.88FAASYY273 pKa = 10.67SFHH276 pKa = 7.36SEE278 pKa = 2.61QWYY281 pKa = 10.02NLRR284 pKa = 11.84SYY286 pKa = 11.18LLRR289 pKa = 11.84LIGLPPHH296 pKa = 7.12IPPNDD301 pKa = 4.74PIRR304 pKa = 11.84VPNRR308 pKa = 11.84VLMLALISRR317 pKa = 11.84LQDD320 pKa = 2.89VYY322 pKa = 11.13VAHH325 pKa = 6.58SPIDD329 pKa = 3.21IVAQQEE335 pKa = 4.61GNGQAGLPDD344 pKa = 3.83AALEE348 pKa = 3.95ALRR351 pKa = 11.84RR352 pKa = 11.84VV353 pKa = 3.65
Molecular weight: 39.53 kDa
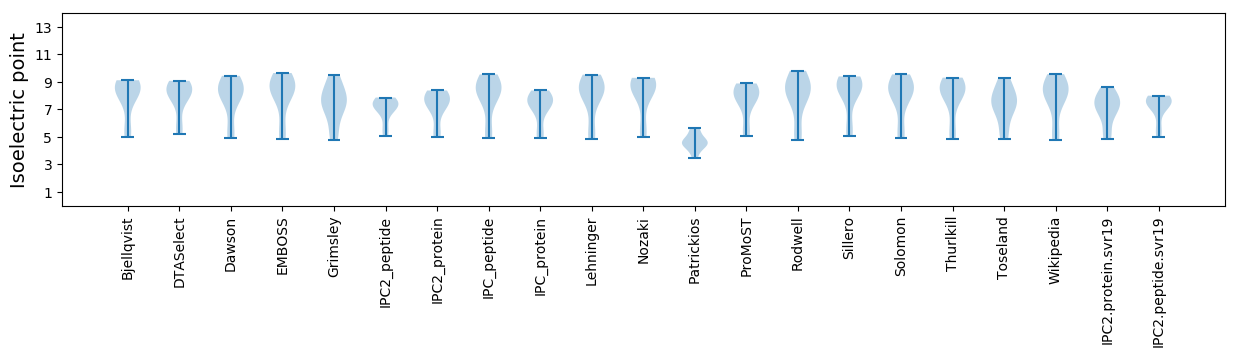
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2Q1D6|Q2Q1D6_9REOV Non-structural protein NS2 OS=Peruvian horse sickness virus OX=356862 PE=4 SV=1
MM1 pKa = 6.65MHH3 pKa = 7.27AALSMTKK10 pKa = 9.91HH11 pKa = 6.0EE12 pKa = 4.62NPISLLTVVPATAPRR27 pKa = 11.84FDD29 pKa = 5.38EE30 pKa = 4.18ITDD33 pKa = 5.05LISSKK38 pKa = 10.73GEE40 pKa = 3.82KK41 pKa = 10.26EE42 pKa = 3.88EE43 pKa = 4.25VKK45 pKa = 9.39ITMSEE50 pKa = 3.93TMNAKK55 pKa = 6.99TTALEE60 pKa = 4.24VLSSALGNGSGTDD73 pKa = 3.21EE74 pKa = 3.68ITRR77 pKa = 11.84RR78 pKa = 11.84EE79 pKa = 3.79RR80 pKa = 11.84SAFGAATQALNEE92 pKa = 4.21EE93 pKa = 4.68EE94 pKa = 3.97NTRR97 pKa = 11.84RR98 pKa = 11.84LKK100 pKa = 11.06VYY102 pKa = 9.6MNEE105 pKa = 4.09QILPKK110 pKa = 10.38LKK112 pKa = 10.44NKK114 pKa = 10.23LMTVTYY120 pKa = 10.11KK121 pKa = 10.54YY122 pKa = 10.39RR123 pKa = 11.84IWLAVEE129 pKa = 4.52IILAIFSMVVFGVMAVNEE147 pKa = 4.39AEE149 pKa = 4.26SEE151 pKa = 4.26INSWLQNKK159 pKa = 10.01LGVKK163 pKa = 9.91VGLSAVSLGCTFALLFASRR182 pKa = 11.84SAGALKK188 pKa = 9.39EE189 pKa = 4.14TVKK192 pKa = 10.53RR193 pKa = 11.84MKK195 pKa = 10.61RR196 pKa = 11.84EE197 pKa = 3.4ITKK200 pKa = 10.29RR201 pKa = 11.84EE202 pKa = 4.14TYY204 pKa = 10.46NHH206 pKa = 6.77IAGTMAAGTEE216 pKa = 3.78RR217 pKa = 11.84RR218 pKa = 11.84VEE220 pKa = 3.97NSEE223 pKa = 4.14SRR225 pKa = 11.84VSNHH229 pKa = 6.48LNAAWINAPGTTNDD243 pKa = 3.16VPGWQLVRR251 pKa = 11.84LKK253 pKa = 10.82EE254 pKa = 4.13GG255 pKa = 3.17
MM1 pKa = 6.65MHH3 pKa = 7.27AALSMTKK10 pKa = 9.91HH11 pKa = 6.0EE12 pKa = 4.62NPISLLTVVPATAPRR27 pKa = 11.84FDD29 pKa = 5.38EE30 pKa = 4.18ITDD33 pKa = 5.05LISSKK38 pKa = 10.73GEE40 pKa = 3.82KK41 pKa = 10.26EE42 pKa = 3.88EE43 pKa = 4.25VKK45 pKa = 9.39ITMSEE50 pKa = 3.93TMNAKK55 pKa = 6.99TTALEE60 pKa = 4.24VLSSALGNGSGTDD73 pKa = 3.21EE74 pKa = 3.68ITRR77 pKa = 11.84RR78 pKa = 11.84EE79 pKa = 3.79RR80 pKa = 11.84SAFGAATQALNEE92 pKa = 4.21EE93 pKa = 4.68EE94 pKa = 3.97NTRR97 pKa = 11.84RR98 pKa = 11.84LKK100 pKa = 11.06VYY102 pKa = 9.6MNEE105 pKa = 4.09QILPKK110 pKa = 10.38LKK112 pKa = 10.44NKK114 pKa = 10.23LMTVTYY120 pKa = 10.11KK121 pKa = 10.54YY122 pKa = 10.39RR123 pKa = 11.84IWLAVEE129 pKa = 4.52IILAIFSMVVFGVMAVNEE147 pKa = 4.39AEE149 pKa = 4.26SEE151 pKa = 4.26INSWLQNKK159 pKa = 10.01LGVKK163 pKa = 9.91VGLSAVSLGCTFALLFASRR182 pKa = 11.84SAGALKK188 pKa = 9.39EE189 pKa = 4.14TVKK192 pKa = 10.53RR193 pKa = 11.84MKK195 pKa = 10.61RR196 pKa = 11.84EE197 pKa = 3.4ITKK200 pKa = 10.29RR201 pKa = 11.84EE202 pKa = 4.14TYY204 pKa = 10.46NHH206 pKa = 6.77IAGTMAAGTEE216 pKa = 3.78RR217 pKa = 11.84RR218 pKa = 11.84VEE220 pKa = 3.97NSEE223 pKa = 4.14SRR225 pKa = 11.84VSNHH229 pKa = 6.48LNAAWINAPGTTNDD243 pKa = 3.16VPGWQLVRR251 pKa = 11.84LKK253 pKa = 10.82EE254 pKa = 4.13GG255 pKa = 3.17
Molecular weight: 28.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6223 |
255 |
1311 |
622.3 |
71.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.448 ± 0.543 | 1.302 ± 0.174 |
5.335 ± 0.44 | 7.312 ± 0.529 |
4.162 ± 0.326 | 5.399 ± 0.307 |
2.105 ± 0.233 | 7.167 ± 0.374 |
6.701 ± 0.812 | 9.417 ± 0.437 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.166 ± 0.24 | 4.853 ± 0.173 |
3.664 ± 0.499 | 3.471 ± 0.293 |
6.299 ± 0.198 | 7.263 ± 0.313 |
5.962 ± 0.36 | 6.492 ± 0.385 |
1.093 ± 0.241 | 3.391 ± 0.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
