
Apis mellifera associated microvirus 34
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
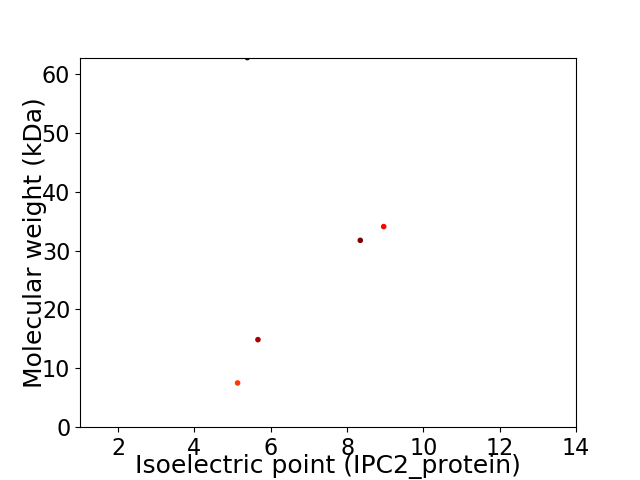
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTD4|A0A3S8UTD4_9VIRU Replication initiator protein OS=Apis mellifera associated microvirus 34 OX=2494763 PE=4 SV=1
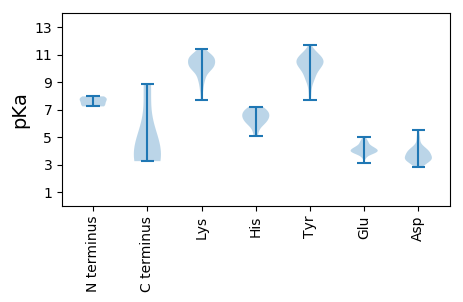
MM1 pKa = 7.23SEE3 pKa = 4.1KK4 pKa = 10.87VKK6 pKa = 10.17TPTKK10 pKa = 10.85AEE12 pKa = 4.0VQQWMQKK19 pKa = 10.71DD20 pKa = 3.06IATAVAFLNMLRR32 pKa = 11.84NDD34 pKa = 3.52QAMLDD39 pKa = 3.81ALADD43 pKa = 3.67VALEE47 pKa = 4.11RR48 pKa = 11.84IKK50 pKa = 10.93AQEE53 pKa = 3.97EE54 pKa = 4.65SKK56 pKa = 10.51QVQPEE61 pKa = 4.16LPLTNGG67 pKa = 3.25
MM1 pKa = 7.23SEE3 pKa = 4.1KK4 pKa = 10.87VKK6 pKa = 10.17TPTKK10 pKa = 10.85AEE12 pKa = 4.0VQQWMQKK19 pKa = 10.71DD20 pKa = 3.06IATAVAFLNMLRR32 pKa = 11.84NDD34 pKa = 3.52QAMLDD39 pKa = 3.81ALADD43 pKa = 3.67VALEE47 pKa = 4.11RR48 pKa = 11.84IKK50 pKa = 10.93AQEE53 pKa = 3.97EE54 pKa = 4.65SKK56 pKa = 10.51QVQPEE61 pKa = 4.16LPLTNGG67 pKa = 3.25
Molecular weight: 7.51 kDa
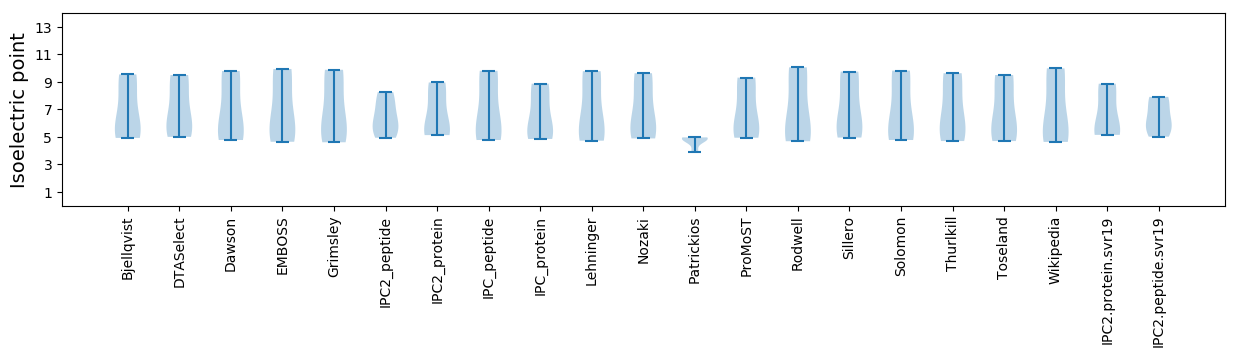
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTE6|A0A3S8UTE6_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 34 OX=2494763 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.25CPSPNFVWPHH12 pKa = 6.3DD13 pKa = 3.77KK14 pKa = 10.91VIAVPCGKK22 pKa = 10.25CLVCLSNKK30 pKa = 9.12RR31 pKa = 11.84HH32 pKa = 6.0DD33 pKa = 3.28WSVRR37 pKa = 11.84LMQEE41 pKa = 4.01YY42 pKa = 9.92KK43 pKa = 10.92VSDD46 pKa = 3.28NAFFVTLTYY55 pKa = 10.43SDD57 pKa = 4.33KK58 pKa = 10.39YY59 pKa = 10.66LPSFGVSKK67 pKa = 10.73RR68 pKa = 11.84HH69 pKa = 5.37LQLFFKK75 pKa = 10.5RR76 pKa = 11.84VRR78 pKa = 11.84KK79 pKa = 8.59EE80 pKa = 3.87CNRR83 pKa = 11.84LRR85 pKa = 11.84YY86 pKa = 9.15YY87 pKa = 11.14AVGEE91 pKa = 4.16YY92 pKa = 10.61GSRR95 pKa = 11.84TGRR98 pKa = 11.84PHH100 pKa = 5.09YY101 pKa = 9.66HH102 pKa = 7.0AIMFNVDD109 pKa = 3.45PKK111 pKa = 11.33VVTDD115 pKa = 4.01CWTLFNKK122 pKa = 7.66VTNKK126 pKa = 10.06KK127 pKa = 9.64EE128 pKa = 4.32PIGLVHH134 pKa = 6.78FGKK137 pKa = 8.82VTEE140 pKa = 4.62ASVQYY145 pKa = 7.68TLKK148 pKa = 11.22YY149 pKa = 9.59IVQKK153 pKa = 11.37NEE155 pKa = 3.69FPKK158 pKa = 10.52EE159 pKa = 3.97LNPSFSLMSRR169 pKa = 11.84GYY171 pKa = 11.0GLGLNYY177 pKa = 10.52LSDD180 pKa = 5.19RR181 pKa = 11.84MLSWHH186 pKa = 7.01RR187 pKa = 11.84DD188 pKa = 3.23DD189 pKa = 5.51DD190 pKa = 3.97RR191 pKa = 11.84VYY193 pKa = 10.48MVVDD197 pKa = 4.1GEE199 pKa = 4.4KK200 pKa = 10.33KK201 pKa = 10.13RR202 pKa = 11.84LPRR205 pKa = 11.84YY206 pKa = 8.58YY207 pKa = 10.19KK208 pKa = 10.31EE209 pKa = 4.76KK210 pKa = 9.25IWPAIKK216 pKa = 10.48QEE218 pKa = 4.9DD219 pKa = 4.71YY220 pKa = 9.97PQGWARR226 pKa = 11.84WTWRR230 pKa = 11.84RR231 pKa = 11.84EE232 pKa = 3.72DD233 pKa = 3.97CFNKK237 pKa = 9.79ARR239 pKa = 11.84KK240 pKa = 8.2EE241 pKa = 3.93AEE243 pKa = 3.45EE244 pKa = 4.05AEE246 pKa = 4.37RR247 pKa = 11.84YY248 pKa = 9.28NVEE251 pKa = 4.03KK252 pKa = 10.8VRR254 pKa = 11.84SEE256 pKa = 4.63GYY258 pKa = 10.4SNPEE262 pKa = 3.5QVITEE267 pKa = 3.88MRR269 pKa = 11.84NAVVNRR275 pKa = 11.84IQSKK279 pKa = 9.19VAFTQKK285 pKa = 10.02VFF287 pKa = 3.35
MM1 pKa = 7.65KK2 pKa = 10.25CPSPNFVWPHH12 pKa = 6.3DD13 pKa = 3.77KK14 pKa = 10.91VIAVPCGKK22 pKa = 10.25CLVCLSNKK30 pKa = 9.12RR31 pKa = 11.84HH32 pKa = 6.0DD33 pKa = 3.28WSVRR37 pKa = 11.84LMQEE41 pKa = 4.01YY42 pKa = 9.92KK43 pKa = 10.92VSDD46 pKa = 3.28NAFFVTLTYY55 pKa = 10.43SDD57 pKa = 4.33KK58 pKa = 10.39YY59 pKa = 10.66LPSFGVSKK67 pKa = 10.73RR68 pKa = 11.84HH69 pKa = 5.37LQLFFKK75 pKa = 10.5RR76 pKa = 11.84VRR78 pKa = 11.84KK79 pKa = 8.59EE80 pKa = 3.87CNRR83 pKa = 11.84LRR85 pKa = 11.84YY86 pKa = 9.15YY87 pKa = 11.14AVGEE91 pKa = 4.16YY92 pKa = 10.61GSRR95 pKa = 11.84TGRR98 pKa = 11.84PHH100 pKa = 5.09YY101 pKa = 9.66HH102 pKa = 7.0AIMFNVDD109 pKa = 3.45PKK111 pKa = 11.33VVTDD115 pKa = 4.01CWTLFNKK122 pKa = 7.66VTNKK126 pKa = 10.06KK127 pKa = 9.64EE128 pKa = 4.32PIGLVHH134 pKa = 6.78FGKK137 pKa = 8.82VTEE140 pKa = 4.62ASVQYY145 pKa = 7.68TLKK148 pKa = 11.22YY149 pKa = 9.59IVQKK153 pKa = 11.37NEE155 pKa = 3.69FPKK158 pKa = 10.52EE159 pKa = 3.97LNPSFSLMSRR169 pKa = 11.84GYY171 pKa = 11.0GLGLNYY177 pKa = 10.52LSDD180 pKa = 5.19RR181 pKa = 11.84MLSWHH186 pKa = 7.01RR187 pKa = 11.84DD188 pKa = 3.23DD189 pKa = 5.51DD190 pKa = 3.97RR191 pKa = 11.84VYY193 pKa = 10.48MVVDD197 pKa = 4.1GEE199 pKa = 4.4KK200 pKa = 10.33KK201 pKa = 10.13RR202 pKa = 11.84LPRR205 pKa = 11.84YY206 pKa = 8.58YY207 pKa = 10.19KK208 pKa = 10.31EE209 pKa = 4.76KK210 pKa = 9.25IWPAIKK216 pKa = 10.48QEE218 pKa = 4.9DD219 pKa = 4.71YY220 pKa = 9.97PQGWARR226 pKa = 11.84WTWRR230 pKa = 11.84RR231 pKa = 11.84EE232 pKa = 3.72DD233 pKa = 3.97CFNKK237 pKa = 9.79ARR239 pKa = 11.84KK240 pKa = 8.2EE241 pKa = 3.93AEE243 pKa = 3.45EE244 pKa = 4.05AEE246 pKa = 4.37RR247 pKa = 11.84YY248 pKa = 9.28NVEE251 pKa = 4.03KK252 pKa = 10.8VRR254 pKa = 11.84SEE256 pKa = 4.63GYY258 pKa = 10.4SNPEE262 pKa = 3.5QVITEE267 pKa = 3.88MRR269 pKa = 11.84NAVVNRR275 pKa = 11.84IQSKK279 pKa = 9.19VAFTQKK285 pKa = 10.02VFF287 pKa = 3.35
Molecular weight: 34.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1317 |
67 |
551 |
263.4 |
30.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.669 ± 1.303 | 0.759 ± 0.403 |
5.695 ± 0.551 | 6.986 ± 1.173 |
4.556 ± 0.444 | 5.695 ± 0.54 |
1.898 ± 0.523 | 3.721 ± 0.308 |
7.137 ± 1.825 | 7.062 ± 0.559 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.797 ± 0.65 | 4.784 ± 0.474 |
4.784 ± 0.707 | 4.328 ± 1.344 |
5.923 ± 0.724 | 6.302 ± 0.856 |
5.239 ± 0.486 | 7.213 ± 0.722 |
2.202 ± 0.275 | 4.252 ± 0.608 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
