
Thermomonospora umbrina
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Thermomonospora
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
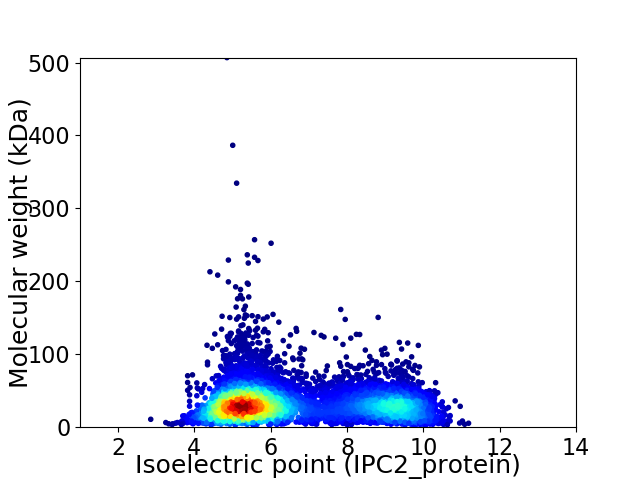
Virtual 2D-PAGE plot for 6565 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D9SSA7|A0A3D9SSA7_9ACTN Uncharacterized protein DUF4232 OS=Thermomonospora umbrina OX=111806 GN=DFJ69_1259 PE=4 SV=1
MM1 pKa = 7.15MMYY4 pKa = 9.98IVVPGLIVVVILVVVLVALGMRR26 pKa = 11.84ANRR29 pKa = 11.84TGADD33 pKa = 3.35EE34 pKa = 5.89DD35 pKa = 4.06DD36 pKa = 4.37WADD39 pKa = 4.46EE40 pKa = 4.56PPQQQPRR47 pKa = 11.84SRR49 pKa = 11.84GRR51 pKa = 11.84RR52 pKa = 11.84AAPPPVDD59 pKa = 3.72EE60 pKa = 5.18PVDD63 pKa = 4.23DD64 pKa = 4.94GYY66 pKa = 11.59EE67 pKa = 3.96DD68 pKa = 4.04HH69 pKa = 6.85GTGAGYY75 pKa = 10.25DD76 pKa = 3.22RR77 pKa = 11.84RR78 pKa = 11.84VAGGPLAAPTGPPAPAQPAGLAGPGPAGAPPGPPAPPPPAPRR120 pKa = 11.84RR121 pKa = 11.84APAGDD126 pKa = 3.89DD127 pKa = 3.53MSDD130 pKa = 3.06EE131 pKa = 4.67MGDD134 pKa = 3.49DD135 pKa = 4.94DD136 pKa = 4.56YY137 pKa = 11.54WATITFDD144 pKa = 3.31KK145 pKa = 11.02PKK147 pKa = 10.48FPWQHH152 pKa = 6.53DD153 pKa = 3.51RR154 pKa = 11.84DD155 pKa = 3.82EE156 pKa = 5.19DD157 pKa = 4.01RR158 pKa = 11.84DD159 pKa = 3.7EE160 pKa = 4.93AADD163 pKa = 3.96PLAAAADD170 pKa = 3.98MAEE173 pKa = 4.68PGAPQQPHH181 pKa = 6.62APAEE185 pKa = 3.98PAMAQPVPAGPMDD198 pKa = 3.43QPGYY202 pKa = 10.55GGDD205 pKa = 3.55SFGSDD210 pKa = 3.12GQYY213 pKa = 11.33GADD216 pKa = 3.13QYY218 pKa = 12.02GAGDD222 pKa = 3.36QYY224 pKa = 11.89GAGDD228 pKa = 3.47QYY230 pKa = 11.78ATGDD234 pKa = 3.43QYY236 pKa = 11.95GVEE239 pKa = 4.39QYY241 pKa = 11.19GADD244 pKa = 3.41QYY246 pKa = 12.02GGGGDD251 pKa = 3.69QYY253 pKa = 11.71ADD255 pKa = 3.32QYY257 pKa = 10.8GDD259 pKa = 3.36QYY261 pKa = 11.82ADD263 pKa = 3.45QPADD267 pKa = 3.18QGDD270 pKa = 4.13RR271 pKa = 11.84YY272 pKa = 10.57AADD275 pKa = 3.51GGRR278 pKa = 11.84YY279 pKa = 8.16PVEE282 pKa = 4.04PAPYY286 pKa = 8.97AAEE289 pKa = 4.27AGPYY293 pKa = 9.53GGEE296 pKa = 4.01PAASPGGYY304 pKa = 8.26GTGPQSVPAPPPSFPGTPEE323 pKa = 3.8PHH325 pKa = 7.27DD326 pKa = 4.34LPIAAHH332 pKa = 5.76FRR334 pKa = 11.84GDD336 pKa = 3.6AGDD339 pKa = 3.98PLATGGNPAGPTRR352 pKa = 11.84QDD354 pKa = 3.5PLDD357 pKa = 4.06YY358 pKa = 9.37ATPPPVSDD366 pKa = 3.98PLGLPVGRR374 pKa = 11.84ADD376 pKa = 4.4DD377 pKa = 4.12RR378 pKa = 11.84SSTGPFAGPGEE389 pKa = 4.46GSFGLPDD396 pKa = 3.9PPSYY400 pKa = 11.05GGAGEE405 pKa = 4.6GAGSTGYY412 pKa = 10.96GPTEE416 pKa = 4.03TTVSFSADD424 pKa = 3.47SPPAGPLHH432 pKa = 6.7GGFGDD437 pKa = 4.11SRR439 pKa = 11.84GLPRR443 pKa = 11.84NEE445 pKa = 3.99PSGAGPGGPPAHH457 pKa = 6.86SAPAAYY463 pKa = 9.49GSPAAPSGQQNDD475 pKa = 3.9TDD477 pKa = 3.84GHH479 pKa = 6.19RR480 pKa = 11.84LPTVDD485 pKa = 5.33EE486 pKa = 4.18LLQRR490 pKa = 11.84IQSDD494 pKa = 3.94RR495 pKa = 11.84QRR497 pKa = 11.84STGPTPVSGGNPLPDD512 pKa = 4.61PLNDD516 pKa = 4.18PLSTSHH522 pKa = 7.1DD523 pKa = 3.34ASYY526 pKa = 11.49GSGGAPWSSSTTGGSQGYY544 pKa = 8.34DD545 pKa = 3.09QPSSGRR551 pKa = 11.84GHH553 pKa = 7.76DD554 pKa = 4.34DD555 pKa = 3.31LSSGQLSGSGLSGGGFSGLGGGGSPGYY582 pKa = 10.02GQGGSGGQGQGYY594 pKa = 10.29ASGEE598 pKa = 4.47GYY600 pKa = 9.92PSAPAYY606 pKa = 10.52GEE608 pKa = 3.92APRR611 pKa = 11.84YY612 pKa = 9.44DD613 pKa = 4.14DD614 pKa = 4.61PLGGGPSSGRR624 pKa = 11.84EE625 pKa = 3.93SFGSFGMGEE634 pKa = 4.43PSTPPASGGSADD646 pKa = 3.58PGRR649 pKa = 11.84YY650 pKa = 9.67GDD652 pKa = 4.46FSGSNYY658 pKa = 10.6GGGDD662 pKa = 3.58PNATQAFGNGYY673 pKa = 10.32FGGQQGGHH681 pKa = 5.35GQADD685 pKa = 3.21QGTPQGEE692 pKa = 4.16QGSGYY697 pKa = 9.7GQQDD701 pKa = 3.21QRR703 pKa = 11.84GGDD706 pKa = 3.42DD707 pKa = 3.49WEE709 pKa = 4.13SHH711 pKa = 5.64RR712 pKa = 11.84DD713 pKa = 3.49YY714 pKa = 11.28RR715 pKa = 11.84RR716 pKa = 2.95
MM1 pKa = 7.15MMYY4 pKa = 9.98IVVPGLIVVVILVVVLVALGMRR26 pKa = 11.84ANRR29 pKa = 11.84TGADD33 pKa = 3.35EE34 pKa = 5.89DD35 pKa = 4.06DD36 pKa = 4.37WADD39 pKa = 4.46EE40 pKa = 4.56PPQQQPRR47 pKa = 11.84SRR49 pKa = 11.84GRR51 pKa = 11.84RR52 pKa = 11.84AAPPPVDD59 pKa = 3.72EE60 pKa = 5.18PVDD63 pKa = 4.23DD64 pKa = 4.94GYY66 pKa = 11.59EE67 pKa = 3.96DD68 pKa = 4.04HH69 pKa = 6.85GTGAGYY75 pKa = 10.25DD76 pKa = 3.22RR77 pKa = 11.84RR78 pKa = 11.84VAGGPLAAPTGPPAPAQPAGLAGPGPAGAPPGPPAPPPPAPRR120 pKa = 11.84RR121 pKa = 11.84APAGDD126 pKa = 3.89DD127 pKa = 3.53MSDD130 pKa = 3.06EE131 pKa = 4.67MGDD134 pKa = 3.49DD135 pKa = 4.94DD136 pKa = 4.56YY137 pKa = 11.54WATITFDD144 pKa = 3.31KK145 pKa = 11.02PKK147 pKa = 10.48FPWQHH152 pKa = 6.53DD153 pKa = 3.51RR154 pKa = 11.84DD155 pKa = 3.82EE156 pKa = 5.19DD157 pKa = 4.01RR158 pKa = 11.84DD159 pKa = 3.7EE160 pKa = 4.93AADD163 pKa = 3.96PLAAAADD170 pKa = 3.98MAEE173 pKa = 4.68PGAPQQPHH181 pKa = 6.62APAEE185 pKa = 3.98PAMAQPVPAGPMDD198 pKa = 3.43QPGYY202 pKa = 10.55GGDD205 pKa = 3.55SFGSDD210 pKa = 3.12GQYY213 pKa = 11.33GADD216 pKa = 3.13QYY218 pKa = 12.02GAGDD222 pKa = 3.36QYY224 pKa = 11.89GAGDD228 pKa = 3.47QYY230 pKa = 11.78ATGDD234 pKa = 3.43QYY236 pKa = 11.95GVEE239 pKa = 4.39QYY241 pKa = 11.19GADD244 pKa = 3.41QYY246 pKa = 12.02GGGGDD251 pKa = 3.69QYY253 pKa = 11.71ADD255 pKa = 3.32QYY257 pKa = 10.8GDD259 pKa = 3.36QYY261 pKa = 11.82ADD263 pKa = 3.45QPADD267 pKa = 3.18QGDD270 pKa = 4.13RR271 pKa = 11.84YY272 pKa = 10.57AADD275 pKa = 3.51GGRR278 pKa = 11.84YY279 pKa = 8.16PVEE282 pKa = 4.04PAPYY286 pKa = 8.97AAEE289 pKa = 4.27AGPYY293 pKa = 9.53GGEE296 pKa = 4.01PAASPGGYY304 pKa = 8.26GTGPQSVPAPPPSFPGTPEE323 pKa = 3.8PHH325 pKa = 7.27DD326 pKa = 4.34LPIAAHH332 pKa = 5.76FRR334 pKa = 11.84GDD336 pKa = 3.6AGDD339 pKa = 3.98PLATGGNPAGPTRR352 pKa = 11.84QDD354 pKa = 3.5PLDD357 pKa = 4.06YY358 pKa = 9.37ATPPPVSDD366 pKa = 3.98PLGLPVGRR374 pKa = 11.84ADD376 pKa = 4.4DD377 pKa = 4.12RR378 pKa = 11.84SSTGPFAGPGEE389 pKa = 4.46GSFGLPDD396 pKa = 3.9PPSYY400 pKa = 11.05GGAGEE405 pKa = 4.6GAGSTGYY412 pKa = 10.96GPTEE416 pKa = 4.03TTVSFSADD424 pKa = 3.47SPPAGPLHH432 pKa = 6.7GGFGDD437 pKa = 4.11SRR439 pKa = 11.84GLPRR443 pKa = 11.84NEE445 pKa = 3.99PSGAGPGGPPAHH457 pKa = 6.86SAPAAYY463 pKa = 9.49GSPAAPSGQQNDD475 pKa = 3.9TDD477 pKa = 3.84GHH479 pKa = 6.19RR480 pKa = 11.84LPTVDD485 pKa = 5.33EE486 pKa = 4.18LLQRR490 pKa = 11.84IQSDD494 pKa = 3.94RR495 pKa = 11.84QRR497 pKa = 11.84STGPTPVSGGNPLPDD512 pKa = 4.61PLNDD516 pKa = 4.18PLSTSHH522 pKa = 7.1DD523 pKa = 3.34ASYY526 pKa = 11.49GSGGAPWSSSTTGGSQGYY544 pKa = 8.34DD545 pKa = 3.09QPSSGRR551 pKa = 11.84GHH553 pKa = 7.76DD554 pKa = 4.34DD555 pKa = 3.31LSSGQLSGSGLSGGGFSGLGGGGSPGYY582 pKa = 10.02GQGGSGGQGQGYY594 pKa = 10.29ASGEE598 pKa = 4.47GYY600 pKa = 9.92PSAPAYY606 pKa = 10.52GEE608 pKa = 3.92APRR611 pKa = 11.84YY612 pKa = 9.44DD613 pKa = 4.14DD614 pKa = 4.61PLGGGPSSGRR624 pKa = 11.84EE625 pKa = 3.93SFGSFGMGEE634 pKa = 4.43PSTPPASGGSADD646 pKa = 3.58PGRR649 pKa = 11.84YY650 pKa = 9.67GDD652 pKa = 4.46FSGSNYY658 pKa = 10.6GGGDD662 pKa = 3.58PNATQAFGNGYY673 pKa = 10.32FGGQQGGHH681 pKa = 5.35GQADD685 pKa = 3.21QGTPQGEE692 pKa = 4.16QGSGYY697 pKa = 9.7GQQDD701 pKa = 3.21QRR703 pKa = 11.84GGDD706 pKa = 3.42DD707 pKa = 3.49WEE709 pKa = 4.13SHH711 pKa = 5.64RR712 pKa = 11.84DD713 pKa = 3.49YY714 pKa = 11.28RR715 pKa = 11.84RR716 pKa = 2.95
Molecular weight: 71.75 kDa
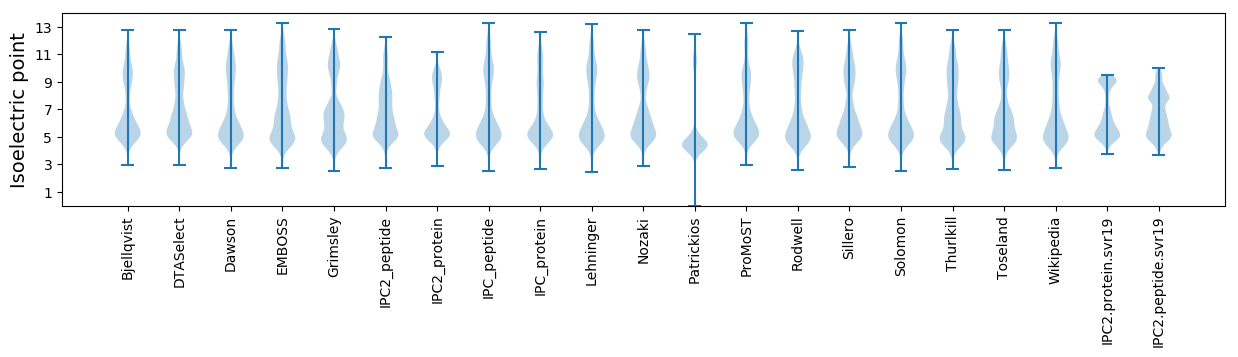
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D9SQA3|A0A3D9SQA3_9ACTN Putative cold-shock DNA-binding protein OS=Thermomonospora umbrina OX=111806 GN=DFJ69_2104 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2119970 |
29 |
4672 |
322.9 |
34.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.193 ± 0.045 | 0.787 ± 0.008 |
6.146 ± 0.023 | 5.729 ± 0.03 |
2.692 ± 0.017 | 9.648 ± 0.039 |
2.275 ± 0.014 | 3.305 ± 0.019 |
1.789 ± 0.023 | 10.283 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.012 | 1.67 ± 0.016 |
6.501 ± 0.035 | 2.401 ± 0.017 |
9.031 ± 0.033 | 4.74 ± 0.024 |
5.855 ± 0.025 | 8.656 ± 0.028 |
1.529 ± 0.012 | 1.939 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
