
SAR11 cluster bacterium PRT-SC02
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Pelagibacterales; Pelagibacteraceae; unclassified Pelagibacteraceae
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
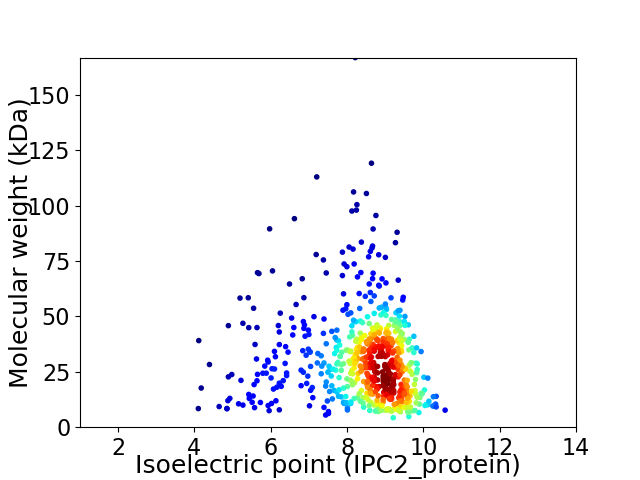
Virtual 2D-PAGE plot for 632 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
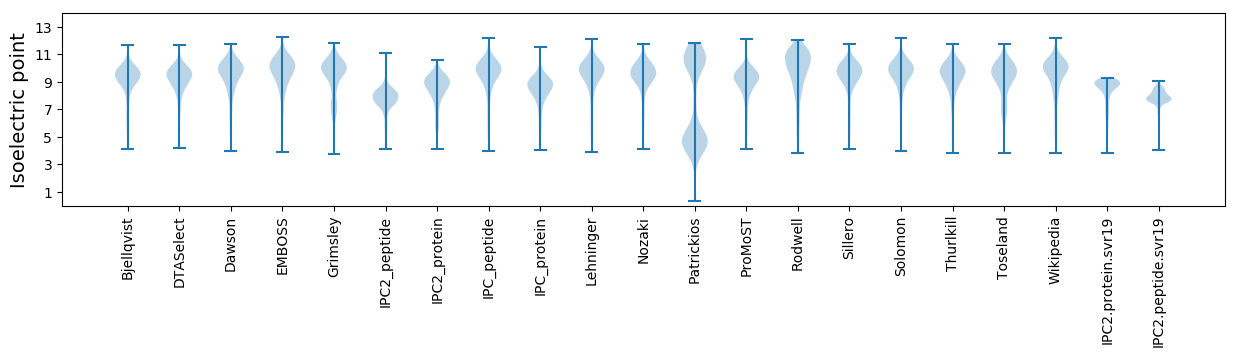
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P9CG28|A0A0P9CG28_9PROT Uncharacterized protein OS=SAR11 cluster bacterium PRT-SC02 OX=1527298 GN=JI56_03950 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.42GKK4 pKa = 10.48DD5 pKa = 3.22MTKK8 pKa = 8.93LTKK11 pKa = 10.32IMLAMIASLSLLSTSYY27 pKa = 10.88AGEE30 pKa = 4.03VTVTGNAEE38 pKa = 3.77ATYY41 pKa = 9.98SISGAKK47 pKa = 9.07TGTGVDD53 pKa = 3.63DD54 pKa = 4.01TDD56 pKa = 3.78TVGGKK61 pKa = 10.5GIGVANEE68 pKa = 3.63IDD70 pKa = 3.84FTATGEE76 pKa = 4.17TDD78 pKa = 4.09NGYY81 pKa = 6.38TWKK84 pKa = 10.68YY85 pKa = 6.68QTQFDD90 pKa = 4.71DD91 pKa = 5.95GSATDD96 pKa = 4.21DD97 pKa = 3.46GRR99 pKa = 11.84LEE101 pKa = 3.9ITSPFGTVGFYY112 pKa = 10.82VSEE115 pKa = 4.5GSLGSHH121 pKa = 6.38YY122 pKa = 10.81GWDD125 pKa = 3.38VSAYY129 pKa = 10.34GAGSDD134 pKa = 4.76QGITTDD140 pKa = 3.8AEE142 pKa = 4.31INGIEE147 pKa = 3.95ASDD150 pKa = 3.81YY151 pKa = 11.83NNIQYY156 pKa = 7.51HH157 pKa = 5.48TPADD161 pKa = 4.02LLPFGIQAKK170 pKa = 9.89VAFAPNGATGQTKK183 pKa = 9.8AASVRR188 pKa = 11.84AQGANTADD196 pKa = 3.27IAEE199 pKa = 4.38ATSYY203 pKa = 10.91QITATPIDD211 pKa = 4.29GLSVGASYY219 pKa = 11.67AEE221 pKa = 4.03ATHH224 pKa = 6.3NGGVTATKK232 pKa = 9.58YY233 pKa = 9.95QPEE236 pKa = 4.2NGSLYY241 pKa = 10.03VKK243 pKa = 10.2YY244 pKa = 10.42AIGPATIGYY253 pKa = 8.09GKK255 pKa = 9.0TYY257 pKa = 9.39IQPRR261 pKa = 11.84VAARR265 pKa = 11.84TADD268 pKa = 3.27TDD270 pKa = 3.78TTNEE274 pKa = 3.36ARR276 pKa = 11.84KK277 pKa = 10.08YY278 pKa = 9.94EE279 pKa = 4.06NTSYY283 pKa = 11.7GLGVSVNDD291 pKa = 3.51QLSVSFTTEE300 pKa = 3.58EE301 pKa = 4.25FEE303 pKa = 4.19LTEE306 pKa = 3.83VDD308 pKa = 3.68VAVATDD314 pKa = 4.1AEE316 pKa = 4.89TSATPVINVDD326 pKa = 4.38TIQAAYY332 pKa = 9.89NVGGMTVAIGHH343 pKa = 6.75AEE345 pKa = 3.89ADD347 pKa = 3.98EE348 pKa = 4.07ISYY351 pKa = 11.2NGDD354 pKa = 2.99NATANDD360 pKa = 3.85EE361 pKa = 4.33HH362 pKa = 7.3DD363 pKa = 3.68TTQTFISVKK372 pKa = 9.52MAFF375 pKa = 3.67
MM1 pKa = 7.5KK2 pKa = 10.42GKK4 pKa = 10.48DD5 pKa = 3.22MTKK8 pKa = 8.93LTKK11 pKa = 10.32IMLAMIASLSLLSTSYY27 pKa = 10.88AGEE30 pKa = 4.03VTVTGNAEE38 pKa = 3.77ATYY41 pKa = 9.98SISGAKK47 pKa = 9.07TGTGVDD53 pKa = 3.63DD54 pKa = 4.01TDD56 pKa = 3.78TVGGKK61 pKa = 10.5GIGVANEE68 pKa = 3.63IDD70 pKa = 3.84FTATGEE76 pKa = 4.17TDD78 pKa = 4.09NGYY81 pKa = 6.38TWKK84 pKa = 10.68YY85 pKa = 6.68QTQFDD90 pKa = 4.71DD91 pKa = 5.95GSATDD96 pKa = 4.21DD97 pKa = 3.46GRR99 pKa = 11.84LEE101 pKa = 3.9ITSPFGTVGFYY112 pKa = 10.82VSEE115 pKa = 4.5GSLGSHH121 pKa = 6.38YY122 pKa = 10.81GWDD125 pKa = 3.38VSAYY129 pKa = 10.34GAGSDD134 pKa = 4.76QGITTDD140 pKa = 3.8AEE142 pKa = 4.31INGIEE147 pKa = 3.95ASDD150 pKa = 3.81YY151 pKa = 11.83NNIQYY156 pKa = 7.51HH157 pKa = 5.48TPADD161 pKa = 4.02LLPFGIQAKK170 pKa = 9.89VAFAPNGATGQTKK183 pKa = 9.8AASVRR188 pKa = 11.84AQGANTADD196 pKa = 3.27IAEE199 pKa = 4.38ATSYY203 pKa = 10.91QITATPIDD211 pKa = 4.29GLSVGASYY219 pKa = 11.67AEE221 pKa = 4.03ATHH224 pKa = 6.3NGGVTATKK232 pKa = 9.58YY233 pKa = 9.95QPEE236 pKa = 4.2NGSLYY241 pKa = 10.03VKK243 pKa = 10.2YY244 pKa = 10.42AIGPATIGYY253 pKa = 8.09GKK255 pKa = 9.0TYY257 pKa = 9.39IQPRR261 pKa = 11.84VAARR265 pKa = 11.84TADD268 pKa = 3.27TDD270 pKa = 3.78TTNEE274 pKa = 3.36ARR276 pKa = 11.84KK277 pKa = 10.08YY278 pKa = 9.94EE279 pKa = 4.06NTSYY283 pKa = 11.7GLGVSVNDD291 pKa = 3.51QLSVSFTTEE300 pKa = 3.58EE301 pKa = 4.25FEE303 pKa = 4.19LTEE306 pKa = 3.83VDD308 pKa = 3.68VAVATDD314 pKa = 4.1AEE316 pKa = 4.89TSATPVINVDD326 pKa = 4.38TIQAAYY332 pKa = 9.89NVGGMTVAIGHH343 pKa = 6.75AEE345 pKa = 3.89ADD347 pKa = 3.98EE348 pKa = 4.07ISYY351 pKa = 11.2NGDD354 pKa = 2.99NATANDD360 pKa = 3.85EE361 pKa = 4.33HH362 pKa = 7.3DD363 pKa = 3.68TTQTFISVKK372 pKa = 9.52MAFF375 pKa = 3.67
Molecular weight: 39.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P9A876|A0A0P9A876_9PROT Phytoene synthase OS=SAR11 cluster bacterium PRT-SC02 OX=1527298 GN=JI56_00990 PE=4 SV=1
SS1 pKa = 6.13TGGRR5 pKa = 11.84NNYY8 pKa = 8.87GRR10 pKa = 11.84ITSRR14 pKa = 11.84HH15 pKa = 5.43RR16 pKa = 11.84GGGAKK21 pKa = 9.36HH22 pKa = 6.82KK23 pKa = 10.65YY24 pKa = 9.96RR25 pKa = 11.84IIDD28 pKa = 3.83FYY30 pKa = 10.74RR31 pKa = 11.84NKK33 pKa = 9.76MNTKK37 pKa = 8.45ATVEE41 pKa = 4.13RR42 pKa = 11.84IEE44 pKa = 4.07YY45 pKa = 10.21DD46 pKa = 3.48PNRR49 pKa = 11.84TAHH52 pKa = 6.71IALINYY58 pKa = 7.4EE59 pKa = 4.31NNIKK63 pKa = 10.66NYY65 pKa = 9.28ILCPQGLKK73 pKa = 10.51KK74 pKa = 10.72GDD76 pKa = 3.83IIEE79 pKa = 4.51SGSNIEE85 pKa = 4.49IKK87 pKa = 10.36IGNNLKK93 pKa = 10.61LRR95 pKa = 11.84DD96 pKa = 3.96IPVGTSVHH104 pKa = 5.73NVEE107 pKa = 4.57LTPGGGGKK115 pKa = 9.91LARR118 pKa = 11.84SAGASVVLSGYY129 pKa = 10.69DD130 pKa = 3.12GDD132 pKa = 4.05YY133 pKa = 11.27AIIKK137 pKa = 9.59LSSGEE142 pKa = 4.05TRR144 pKa = 11.84KK145 pKa = 9.66IRR147 pKa = 11.84SDD149 pKa = 3.45CSATIGVVSNPDD161 pKa = 3.14QKK163 pKa = 11.12NIKK166 pKa = 9.04IGKK169 pKa = 8.76AGRR172 pKa = 11.84NRR174 pKa = 11.84WRR176 pKa = 11.84GIRR179 pKa = 11.84PQSRR183 pKa = 11.84GVAMNPVDD191 pKa = 3.85HH192 pKa = 6.82PHH194 pKa = 6.77GGGEE198 pKa = 4.38GKK200 pKa = 9.0TSGGRR205 pKa = 11.84SPVSPWGQSAKK216 pKa = 10.4GLKK219 pKa = 7.63TRR221 pKa = 11.84KK222 pKa = 9.09PKK224 pKa = 10.68RR225 pKa = 11.84SDD227 pKa = 2.68KK228 pKa = 10.96FIISRR233 pKa = 11.84RR234 pKa = 11.84KK235 pKa = 8.88KK236 pKa = 10.22RR237 pKa = 11.84KK238 pKa = 8.99
SS1 pKa = 6.13TGGRR5 pKa = 11.84NNYY8 pKa = 8.87GRR10 pKa = 11.84ITSRR14 pKa = 11.84HH15 pKa = 5.43RR16 pKa = 11.84GGGAKK21 pKa = 9.36HH22 pKa = 6.82KK23 pKa = 10.65YY24 pKa = 9.96RR25 pKa = 11.84IIDD28 pKa = 3.83FYY30 pKa = 10.74RR31 pKa = 11.84NKK33 pKa = 9.76MNTKK37 pKa = 8.45ATVEE41 pKa = 4.13RR42 pKa = 11.84IEE44 pKa = 4.07YY45 pKa = 10.21DD46 pKa = 3.48PNRR49 pKa = 11.84TAHH52 pKa = 6.71IALINYY58 pKa = 7.4EE59 pKa = 4.31NNIKK63 pKa = 10.66NYY65 pKa = 9.28ILCPQGLKK73 pKa = 10.51KK74 pKa = 10.72GDD76 pKa = 3.83IIEE79 pKa = 4.51SGSNIEE85 pKa = 4.49IKK87 pKa = 10.36IGNNLKK93 pKa = 10.61LRR95 pKa = 11.84DD96 pKa = 3.96IPVGTSVHH104 pKa = 5.73NVEE107 pKa = 4.57LTPGGGGKK115 pKa = 9.91LARR118 pKa = 11.84SAGASVVLSGYY129 pKa = 10.69DD130 pKa = 3.12GDD132 pKa = 4.05YY133 pKa = 11.27AIIKK137 pKa = 9.59LSSGEE142 pKa = 4.05TRR144 pKa = 11.84KK145 pKa = 9.66IRR147 pKa = 11.84SDD149 pKa = 3.45CSATIGVVSNPDD161 pKa = 3.14QKK163 pKa = 11.12NIKK166 pKa = 9.04IGKK169 pKa = 8.76AGRR172 pKa = 11.84NRR174 pKa = 11.84WRR176 pKa = 11.84GIRR179 pKa = 11.84PQSRR183 pKa = 11.84GVAMNPVDD191 pKa = 3.85HH192 pKa = 6.82PHH194 pKa = 6.77GGGEE198 pKa = 4.38GKK200 pKa = 9.0TSGGRR205 pKa = 11.84SPVSPWGQSAKK216 pKa = 10.4GLKK219 pKa = 7.63TRR221 pKa = 11.84KK222 pKa = 9.09PKK224 pKa = 10.68RR225 pKa = 11.84SDD227 pKa = 2.68KK228 pKa = 10.96FIISRR233 pKa = 11.84RR234 pKa = 11.84KK235 pKa = 8.88KK236 pKa = 10.22RR237 pKa = 11.84KK238 pKa = 8.99
Molecular weight: 26.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
174364 |
36 |
1499 |
275.9 |
31.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.154 ± 0.102 | 0.979 ± 0.031 |
4.928 ± 0.073 | 5.74 ± 0.096 |
5.516 ± 0.097 | 5.959 ± 0.09 |
1.49 ± 0.041 | 9.953 ± 0.11 |
11.799 ± 0.154 | 9.472 ± 0.101 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.018 ± 0.04 | 6.807 ± 0.107 |
3.044 ± 0.046 | 2.552 ± 0.049 |
3.367 ± 0.063 | 6.979 ± 0.076 |
4.486 ± 0.054 | 5.454 ± 0.069 |
0.884 ± 0.035 | 3.419 ± 0.06 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
