
Legionella worsleiensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales; Legionellaceae; Legionella
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
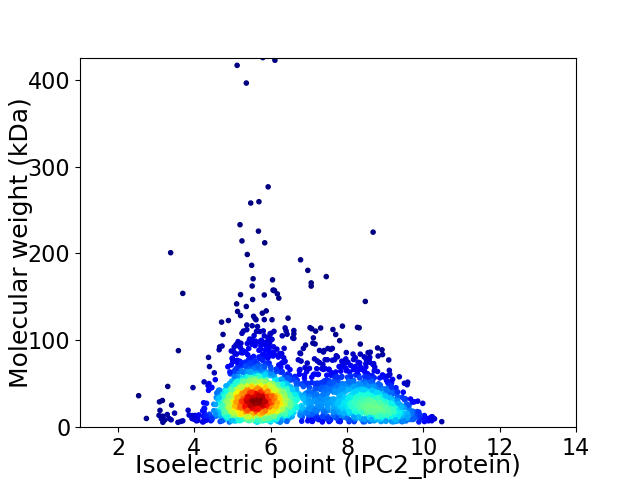
Virtual 2D-PAGE plot for 2614 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W1A3H8|A0A0W1A3H8_9GAMM SidA protein subsrate of the Dot/Icm transport system OS=Legionella worsleiensis OX=45076 GN=sidA PE=4 SV=1
MM1 pKa = 6.95VANGSVVLNADD12 pKa = 3.52GTLSFTPAADD22 pKa = 3.91FNGNASFTYY31 pKa = 9.12TVTSAGLTEE40 pKa = 4.13TATVNVTVNPVNEE53 pKa = 4.1APINSTPGAQTLSEE67 pKa = 4.14DD68 pKa = 3.28ATRR71 pKa = 11.84VFSSANGNALSVTDD85 pKa = 3.72IDD87 pKa = 4.17SAVVTTTISVLHH99 pKa = 6.42GSLTALATAGVTISNNGTGSVTLSGAPAAINTALNGLIYY138 pKa = 10.77SPVADD143 pKa = 3.58YY144 pKa = 11.0HH145 pKa = 7.57GSDD148 pKa = 3.96TLTMSTTDD156 pKa = 3.71GSLLDD161 pKa = 4.09SDD163 pKa = 4.54TVGITINPVVDD174 pKa = 3.58IANDD178 pKa = 4.09TIVTNEE184 pKa = 3.82DD185 pKa = 3.22TAVIVNVNANDD196 pKa = 3.59SFEE199 pKa = 4.44NAGHH203 pKa = 7.32TITAINGTTIAVGGTVAVADD223 pKa = 4.25GSVLLNANGTLTFTPSANYY242 pKa = 10.5NGTTNFTYY250 pKa = 10.0TVTSGGVTEE259 pKa = 4.24TASVNVTVNAVNDD272 pKa = 3.85APQGQNNLLSVNEE285 pKa = 4.21DD286 pKa = 3.59TPIVLSAANFNFTDD300 pKa = 4.08TDD302 pKa = 3.61DD303 pKa = 3.93TPANTFAGVRR313 pKa = 11.84ITTLPANGTLTLNGVAVVAGQTISIANINAGLLVFTPTADD353 pKa = 3.54YY354 pKa = 11.06SGQTNFTFQVEE365 pKa = 4.73DD366 pKa = 3.29NGGTANGGVNLDD378 pKa = 3.46PTPNVMTVDD387 pKa = 3.72VASVADD393 pKa = 3.9TPVVFAHH400 pKa = 6.97LSLPTLVAGTAVIAANFSNNSNEE423 pKa = 4.09NNLSNWSRR431 pKa = 11.84VALFSNTFAEE441 pKa = 4.38IQGGSGKK448 pKa = 8.94TNTEE452 pKa = 3.91ANLFSQIVWAGEE464 pKa = 3.93TGTNATEE471 pKa = 3.82VSNFNNRR478 pKa = 11.84WTKK481 pKa = 9.88PSGTIIYY488 pKa = 8.56NANEE492 pKa = 4.18VADD495 pKa = 4.39DD496 pKa = 3.89AQGILVLNSNQLTVAEE512 pKa = 4.33RR513 pKa = 11.84ALTNYY518 pKa = 9.3VLSVDD523 pKa = 4.06MFADD527 pKa = 3.89ADD529 pKa = 3.91SPRR532 pKa = 11.84ANGTGIVFGYY542 pKa = 10.51QNASNYY548 pKa = 10.17FLVRR552 pKa = 11.84WEE554 pKa = 4.36NPSIEE559 pKa = 4.07YY560 pKa = 10.77SPGGEE565 pKa = 4.09FFNVHH570 pKa = 6.38PGQYY574 pKa = 9.65QEE576 pKa = 4.07LTLVQVVGNVPIDD589 pKa = 3.59LARR592 pKa = 11.84TTFNADD598 pKa = 2.32DD599 pKa = 3.84WFNVVINVSNLGISVSASDD618 pKa = 3.96MSNPSITANFDD629 pKa = 3.67YY630 pKa = 10.52IYY632 pKa = 10.79GSVSGAAAAAPALRR646 pKa = 11.84TIGLYY651 pKa = 9.93TYY653 pKa = 10.71DD654 pKa = 4.11NDD656 pKa = 5.9SAVQWDD662 pKa = 4.67NINVNQPSSYY672 pKa = 10.72QYY674 pKa = 9.96TLQTEE679 pKa = 5.19AYY681 pKa = 9.64LKK683 pKa = 11.08DD684 pKa = 3.56NDD686 pKa = 4.05GSEE689 pKa = 4.26TLGNITLAGIPAGVTLLDD707 pKa = 3.62VVTNTSIPVNAGSATIIPGNDD728 pKa = 2.71VRR730 pKa = 11.84LVSSTALTTAQINAITATVSSVEE753 pKa = 4.13SSNGSTASQTTSVQLEE769 pKa = 4.47LNGGYY774 pKa = 10.52APEE777 pKa = 4.54TFTGTNASEE786 pKa = 3.99WLEE789 pKa = 4.06GDD791 pKa = 3.59SSNDD795 pKa = 2.9ILNAGGGNDD804 pKa = 4.08TIIGGLGNDD813 pKa = 4.51TITTGSGSDD822 pKa = 3.56QIVLLKK828 pKa = 10.66GQGGATAGAAPTDD841 pKa = 3.98TITDD845 pKa = 3.75FTVNVDD851 pKa = 3.8TIVIKK856 pKa = 9.29GTNIIGVAVSTPVSNTYY873 pKa = 9.87TITVNYY879 pKa = 9.98SGGAPTEE886 pKa = 4.19YY887 pKa = 10.66FKK889 pKa = 10.96VTLSNGAVLNDD900 pKa = 3.82SGDD903 pKa = 3.36THH905 pKa = 6.09LTGVVVSGGTATIDD919 pKa = 3.44GTIVGAILYY928 pKa = 10.51LDD930 pKa = 3.64INSNNQEE937 pKa = 4.14DD938 pKa = 3.64EE939 pKa = 4.28GEE941 pKa = 4.1RR942 pKa = 11.84LGITDD947 pKa = 3.25QYY949 pKa = 11.69GHH951 pKa = 5.83VEE953 pKa = 4.05WVLDD957 pKa = 4.07LAKK960 pKa = 10.59LDD962 pKa = 3.95VNGDD966 pKa = 3.38GQYY969 pKa = 11.55VLGEE973 pKa = 3.89ARR975 pKa = 11.84AVQTGGFDD983 pKa = 3.3IDD985 pKa = 3.62TGLSYY990 pKa = 10.52EE991 pKa = 4.32INLFGPVGSAIVSPLTSLLQAQLEE1015 pKa = 4.6SGFDD1019 pKa = 3.63YY1020 pKa = 9.89QTANANIVARR1030 pKa = 11.84LGLPEE1035 pKa = 4.12GTEE1038 pKa = 4.5LISLNPIMGSHH1049 pKa = 7.52DD1050 pKa = 3.66VLAQNAAVMTAAVQFAEE1067 pKa = 4.37IAAIRR1072 pKa = 11.84YY1073 pKa = 7.3STDD1076 pKa = 2.98EE1077 pKa = 3.98GHH1079 pKa = 6.83VSFSVFEE1086 pKa = 5.04AISKK1090 pKa = 10.58ALLEE1094 pKa = 4.35LPEE1097 pKa = 4.26GVIADD1102 pKa = 4.61FSDD1105 pKa = 3.54KK1106 pKa = 11.39AFLQNIASHH1115 pKa = 6.97LDD1117 pKa = 3.52LGDD1120 pKa = 3.45MASPEE1125 pKa = 4.48VIAFMAASQQALQSSMDD1142 pKa = 3.74TLVPGQNALAAISEE1156 pKa = 4.54VQHH1159 pKa = 5.88LTQGSYY1165 pKa = 10.62AQIIEE1170 pKa = 4.73HH1171 pKa = 5.39YY1172 pKa = 8.84TNGYY1176 pKa = 9.1LPAYY1180 pKa = 8.92LLSDD1184 pKa = 3.52ISVIILAFSSSIISYY1199 pKa = 10.98DD1200 pKa = 3.43EE1201 pKa = 4.14LKK1203 pKa = 11.11GFNDD1207 pKa = 3.81QFNTLVNQSAIMLSDD1222 pKa = 3.56GSEE1225 pKa = 4.05SSNKK1229 pKa = 9.93DD1230 pKa = 3.48DD1231 pKa = 5.06FVHH1234 pKa = 6.6EE1235 pKa = 4.44LDD1237 pKa = 3.72SVVNNFVLAHH1247 pKa = 6.18GLDD1250 pKa = 3.7VSHH1253 pKa = 6.66ATDD1256 pKa = 4.1GQQSLDD1262 pKa = 3.91DD1263 pKa = 3.48QHH1265 pKa = 6.67MNALINQSVEE1275 pKa = 4.06DD1276 pKa = 4.4GNVLAHH1282 pKa = 6.08EE1283 pKa = 4.64VNNHH1287 pKa = 5.98EE1288 pKa = 5.08IITDD1292 pKa = 4.31LSSSIGGMEE1301 pKa = 3.9LHH1303 pKa = 6.68GSSGNDD1309 pKa = 3.45FIFGGEE1315 pKa = 4.25GNDD1318 pKa = 4.56LILGGEE1324 pKa = 4.28GDD1326 pKa = 3.74DD1327 pKa = 4.61RR1328 pKa = 11.84LYY1330 pKa = 11.22GGQGDD1335 pKa = 4.17DD1336 pKa = 4.85SILGGLGDD1344 pKa = 4.32DD1345 pKa = 4.58VIHH1348 pKa = 7.0GGMGNDD1354 pKa = 3.74LMTGGGGHH1362 pKa = 7.59DD1363 pKa = 2.59IFVFEE1368 pKa = 4.64KK1369 pKa = 10.79ADD1371 pKa = 3.41IGEE1374 pKa = 4.32EE1375 pKa = 3.93PAIDD1379 pKa = 4.12TITDD1383 pKa = 4.41FKK1385 pKa = 11.37LGIAGDD1391 pKa = 3.82NSGDD1395 pKa = 3.91KK1396 pKa = 9.88STLDD1400 pKa = 3.28LTDD1403 pKa = 4.14LFKK1406 pKa = 11.2EE1407 pKa = 4.41STLSSDD1413 pKa = 4.49SLDD1416 pKa = 3.38TLLQISTIHH1425 pKa = 6.46NEE1427 pKa = 4.12STNQTDD1433 pKa = 3.86TVIKK1437 pKa = 9.85TDD1439 pKa = 3.51PTGNAQFNNSPEE1451 pKa = 4.2TIILSGVDD1459 pKa = 3.17VASAYY1464 pKa = 7.66GTTNSVEE1471 pKa = 4.37LVQHH1475 pKa = 5.92MLAANVLVMGHH1486 pKa = 6.38
MM1 pKa = 6.95VANGSVVLNADD12 pKa = 3.52GTLSFTPAADD22 pKa = 3.91FNGNASFTYY31 pKa = 9.12TVTSAGLTEE40 pKa = 4.13TATVNVTVNPVNEE53 pKa = 4.1APINSTPGAQTLSEE67 pKa = 4.14DD68 pKa = 3.28ATRR71 pKa = 11.84VFSSANGNALSVTDD85 pKa = 3.72IDD87 pKa = 4.17SAVVTTTISVLHH99 pKa = 6.42GSLTALATAGVTISNNGTGSVTLSGAPAAINTALNGLIYY138 pKa = 10.77SPVADD143 pKa = 3.58YY144 pKa = 11.0HH145 pKa = 7.57GSDD148 pKa = 3.96TLTMSTTDD156 pKa = 3.71GSLLDD161 pKa = 4.09SDD163 pKa = 4.54TVGITINPVVDD174 pKa = 3.58IANDD178 pKa = 4.09TIVTNEE184 pKa = 3.82DD185 pKa = 3.22TAVIVNVNANDD196 pKa = 3.59SFEE199 pKa = 4.44NAGHH203 pKa = 7.32TITAINGTTIAVGGTVAVADD223 pKa = 4.25GSVLLNANGTLTFTPSANYY242 pKa = 10.5NGTTNFTYY250 pKa = 10.0TVTSGGVTEE259 pKa = 4.24TASVNVTVNAVNDD272 pKa = 3.85APQGQNNLLSVNEE285 pKa = 4.21DD286 pKa = 3.59TPIVLSAANFNFTDD300 pKa = 4.08TDD302 pKa = 3.61DD303 pKa = 3.93TPANTFAGVRR313 pKa = 11.84ITTLPANGTLTLNGVAVVAGQTISIANINAGLLVFTPTADD353 pKa = 3.54YY354 pKa = 11.06SGQTNFTFQVEE365 pKa = 4.73DD366 pKa = 3.29NGGTANGGVNLDD378 pKa = 3.46PTPNVMTVDD387 pKa = 3.72VASVADD393 pKa = 3.9TPVVFAHH400 pKa = 6.97LSLPTLVAGTAVIAANFSNNSNEE423 pKa = 4.09NNLSNWSRR431 pKa = 11.84VALFSNTFAEE441 pKa = 4.38IQGGSGKK448 pKa = 8.94TNTEE452 pKa = 3.91ANLFSQIVWAGEE464 pKa = 3.93TGTNATEE471 pKa = 3.82VSNFNNRR478 pKa = 11.84WTKK481 pKa = 9.88PSGTIIYY488 pKa = 8.56NANEE492 pKa = 4.18VADD495 pKa = 4.39DD496 pKa = 3.89AQGILVLNSNQLTVAEE512 pKa = 4.33RR513 pKa = 11.84ALTNYY518 pKa = 9.3VLSVDD523 pKa = 4.06MFADD527 pKa = 3.89ADD529 pKa = 3.91SPRR532 pKa = 11.84ANGTGIVFGYY542 pKa = 10.51QNASNYY548 pKa = 10.17FLVRR552 pKa = 11.84WEE554 pKa = 4.36NPSIEE559 pKa = 4.07YY560 pKa = 10.77SPGGEE565 pKa = 4.09FFNVHH570 pKa = 6.38PGQYY574 pKa = 9.65QEE576 pKa = 4.07LTLVQVVGNVPIDD589 pKa = 3.59LARR592 pKa = 11.84TTFNADD598 pKa = 2.32DD599 pKa = 3.84WFNVVINVSNLGISVSASDD618 pKa = 3.96MSNPSITANFDD629 pKa = 3.67YY630 pKa = 10.52IYY632 pKa = 10.79GSVSGAAAAAPALRR646 pKa = 11.84TIGLYY651 pKa = 9.93TYY653 pKa = 10.71DD654 pKa = 4.11NDD656 pKa = 5.9SAVQWDD662 pKa = 4.67NINVNQPSSYY672 pKa = 10.72QYY674 pKa = 9.96TLQTEE679 pKa = 5.19AYY681 pKa = 9.64LKK683 pKa = 11.08DD684 pKa = 3.56NDD686 pKa = 4.05GSEE689 pKa = 4.26TLGNITLAGIPAGVTLLDD707 pKa = 3.62VVTNTSIPVNAGSATIIPGNDD728 pKa = 2.71VRR730 pKa = 11.84LVSSTALTTAQINAITATVSSVEE753 pKa = 4.13SSNGSTASQTTSVQLEE769 pKa = 4.47LNGGYY774 pKa = 10.52APEE777 pKa = 4.54TFTGTNASEE786 pKa = 3.99WLEE789 pKa = 4.06GDD791 pKa = 3.59SSNDD795 pKa = 2.9ILNAGGGNDD804 pKa = 4.08TIIGGLGNDD813 pKa = 4.51TITTGSGSDD822 pKa = 3.56QIVLLKK828 pKa = 10.66GQGGATAGAAPTDD841 pKa = 3.98TITDD845 pKa = 3.75FTVNVDD851 pKa = 3.8TIVIKK856 pKa = 9.29GTNIIGVAVSTPVSNTYY873 pKa = 9.87TITVNYY879 pKa = 9.98SGGAPTEE886 pKa = 4.19YY887 pKa = 10.66FKK889 pKa = 10.96VTLSNGAVLNDD900 pKa = 3.82SGDD903 pKa = 3.36THH905 pKa = 6.09LTGVVVSGGTATIDD919 pKa = 3.44GTIVGAILYY928 pKa = 10.51LDD930 pKa = 3.64INSNNQEE937 pKa = 4.14DD938 pKa = 3.64EE939 pKa = 4.28GEE941 pKa = 4.1RR942 pKa = 11.84LGITDD947 pKa = 3.25QYY949 pKa = 11.69GHH951 pKa = 5.83VEE953 pKa = 4.05WVLDD957 pKa = 4.07LAKK960 pKa = 10.59LDD962 pKa = 3.95VNGDD966 pKa = 3.38GQYY969 pKa = 11.55VLGEE973 pKa = 3.89ARR975 pKa = 11.84AVQTGGFDD983 pKa = 3.3IDD985 pKa = 3.62TGLSYY990 pKa = 10.52EE991 pKa = 4.32INLFGPVGSAIVSPLTSLLQAQLEE1015 pKa = 4.6SGFDD1019 pKa = 3.63YY1020 pKa = 9.89QTANANIVARR1030 pKa = 11.84LGLPEE1035 pKa = 4.12GTEE1038 pKa = 4.5LISLNPIMGSHH1049 pKa = 7.52DD1050 pKa = 3.66VLAQNAAVMTAAVQFAEE1067 pKa = 4.37IAAIRR1072 pKa = 11.84YY1073 pKa = 7.3STDD1076 pKa = 2.98EE1077 pKa = 3.98GHH1079 pKa = 6.83VSFSVFEE1086 pKa = 5.04AISKK1090 pKa = 10.58ALLEE1094 pKa = 4.35LPEE1097 pKa = 4.26GVIADD1102 pKa = 4.61FSDD1105 pKa = 3.54KK1106 pKa = 11.39AFLQNIASHH1115 pKa = 6.97LDD1117 pKa = 3.52LGDD1120 pKa = 3.45MASPEE1125 pKa = 4.48VIAFMAASQQALQSSMDD1142 pKa = 3.74TLVPGQNALAAISEE1156 pKa = 4.54VQHH1159 pKa = 5.88LTQGSYY1165 pKa = 10.62AQIIEE1170 pKa = 4.73HH1171 pKa = 5.39YY1172 pKa = 8.84TNGYY1176 pKa = 9.1LPAYY1180 pKa = 8.92LLSDD1184 pKa = 3.52ISVIILAFSSSIISYY1199 pKa = 10.98DD1200 pKa = 3.43EE1201 pKa = 4.14LKK1203 pKa = 11.11GFNDD1207 pKa = 3.81QFNTLVNQSAIMLSDD1222 pKa = 3.56GSEE1225 pKa = 4.05SSNKK1229 pKa = 9.93DD1230 pKa = 3.48DD1231 pKa = 5.06FVHH1234 pKa = 6.6EE1235 pKa = 4.44LDD1237 pKa = 3.72SVVNNFVLAHH1247 pKa = 6.18GLDD1250 pKa = 3.7VSHH1253 pKa = 6.66ATDD1256 pKa = 4.1GQQSLDD1262 pKa = 3.91DD1263 pKa = 3.48QHH1265 pKa = 6.67MNALINQSVEE1275 pKa = 4.06DD1276 pKa = 4.4GNVLAHH1282 pKa = 6.08EE1283 pKa = 4.64VNNHH1287 pKa = 5.98EE1288 pKa = 5.08IITDD1292 pKa = 4.31LSSSIGGMEE1301 pKa = 3.9LHH1303 pKa = 6.68GSSGNDD1309 pKa = 3.45FIFGGEE1315 pKa = 4.25GNDD1318 pKa = 4.56LILGGEE1324 pKa = 4.28GDD1326 pKa = 3.74DD1327 pKa = 4.61RR1328 pKa = 11.84LYY1330 pKa = 11.22GGQGDD1335 pKa = 4.17DD1336 pKa = 4.85SILGGLGDD1344 pKa = 4.32DD1345 pKa = 4.58VIHH1348 pKa = 7.0GGMGNDD1354 pKa = 3.74LMTGGGGHH1362 pKa = 7.59DD1363 pKa = 2.59IFVFEE1368 pKa = 4.64KK1369 pKa = 10.79ADD1371 pKa = 3.41IGEE1374 pKa = 4.32EE1375 pKa = 3.93PAIDD1379 pKa = 4.12TITDD1383 pKa = 4.41FKK1385 pKa = 11.37LGIAGDD1391 pKa = 3.82NSGDD1395 pKa = 3.91KK1396 pKa = 9.88STLDD1400 pKa = 3.28LTDD1403 pKa = 4.14LFKK1406 pKa = 11.2EE1407 pKa = 4.41STLSSDD1413 pKa = 4.49SLDD1416 pKa = 3.38TLLQISTIHH1425 pKa = 6.46NEE1427 pKa = 4.12STNQTDD1433 pKa = 3.86TVIKK1437 pKa = 9.85TDD1439 pKa = 3.51PTGNAQFNNSPEE1451 pKa = 4.2TIILSGVDD1459 pKa = 3.17VASAYY1464 pKa = 7.66GTTNSVEE1471 pKa = 4.37LVQHH1475 pKa = 5.92MLAANVLVMGHH1486 pKa = 6.38
Molecular weight: 154.03 kDa
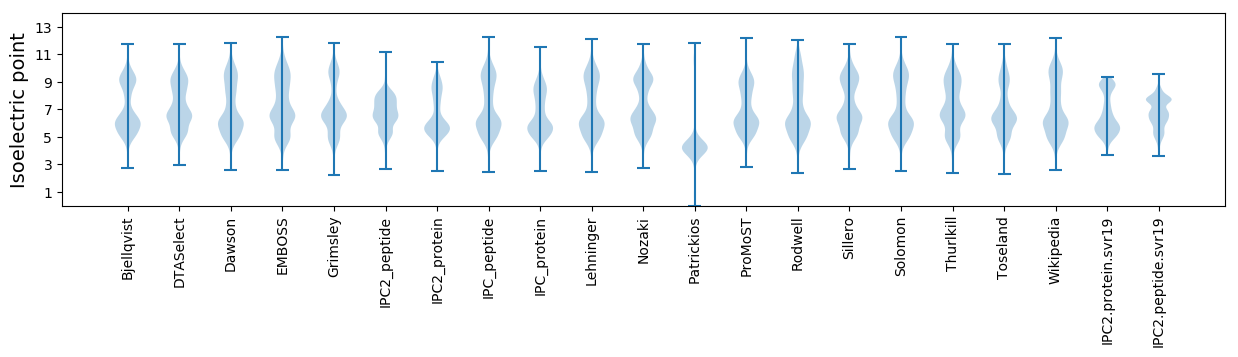
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W1AEJ3|A0A0W1AEJ3_9GAMM Substrate of the Dot/Icm secretion system OS=Legionella worsleiensis OX=45076 GN=pieE_1 PE=4 SV=1
MM1 pKa = 7.82SINTRR6 pKa = 11.84LKK8 pKa = 10.34SIAVLLVSAMLTGCVVAVVAGAAAGMVYY36 pKa = 10.48DD37 pKa = 4.3RR38 pKa = 11.84RR39 pKa = 11.84SVTTIEE45 pKa = 3.47ADD47 pKa = 2.92ARR49 pKa = 11.84LFHH52 pKa = 6.67VIHH55 pKa = 6.77KK56 pKa = 9.53EE57 pKa = 3.83IVTDD61 pKa = 3.79PQFRR65 pKa = 11.84NSRR68 pKa = 11.84ILVTSFNRR76 pKa = 11.84VVLLVGQTPTASLRR90 pKa = 11.84VVAEE94 pKa = 4.26RR95 pKa = 11.84VAQNAPGVRR104 pKa = 11.84RR105 pKa = 11.84VYY107 pKa = 11.17DD108 pKa = 4.65EE109 pKa = 4.05ITVDD113 pKa = 3.62NPIPLTQRR121 pKa = 11.84SQDD124 pKa = 3.53SLITGQVRR132 pKa = 11.84SNMLTKK138 pKa = 10.47KK139 pKa = 9.98GLEE142 pKa = 4.04SGSIRR147 pKa = 11.84IVTEE151 pKa = 3.49NGVVYY156 pKa = 10.59LMGIVTPEE164 pKa = 3.86QASLAVDD171 pKa = 3.58VARR174 pKa = 11.84RR175 pKa = 11.84INGVRR180 pKa = 11.84KK181 pKa = 9.14VVKK184 pKa = 9.21IFQYY188 pKa = 10.51IRR190 pKa = 3.21
MM1 pKa = 7.82SINTRR6 pKa = 11.84LKK8 pKa = 10.34SIAVLLVSAMLTGCVVAVVAGAAAGMVYY36 pKa = 10.48DD37 pKa = 4.3RR38 pKa = 11.84RR39 pKa = 11.84SVTTIEE45 pKa = 3.47ADD47 pKa = 2.92ARR49 pKa = 11.84LFHH52 pKa = 6.67VIHH55 pKa = 6.77KK56 pKa = 9.53EE57 pKa = 3.83IVTDD61 pKa = 3.79PQFRR65 pKa = 11.84NSRR68 pKa = 11.84ILVTSFNRR76 pKa = 11.84VVLLVGQTPTASLRR90 pKa = 11.84VVAEE94 pKa = 4.26RR95 pKa = 11.84VAQNAPGVRR104 pKa = 11.84RR105 pKa = 11.84VYY107 pKa = 11.17DD108 pKa = 4.65EE109 pKa = 4.05ITVDD113 pKa = 3.62NPIPLTQRR121 pKa = 11.84SQDD124 pKa = 3.53SLITGQVRR132 pKa = 11.84SNMLTKK138 pKa = 10.47KK139 pKa = 9.98GLEE142 pKa = 4.04SGSIRR147 pKa = 11.84IVTEE151 pKa = 3.49NGVVYY156 pKa = 10.59LMGIVTPEE164 pKa = 3.86QASLAVDD171 pKa = 3.58VARR174 pKa = 11.84RR175 pKa = 11.84INGVRR180 pKa = 11.84KK181 pKa = 9.14VVKK184 pKa = 9.21IFQYY188 pKa = 10.51IRR190 pKa = 3.21
Molecular weight: 20.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
893477 |
51 |
3762 |
341.8 |
38.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.835 ± 0.05 | 1.261 ± 0.018 |
4.99 ± 0.037 | 5.712 ± 0.047 |
4.373 ± 0.039 | 6.003 ± 0.053 |
2.713 ± 0.023 | 7.085 ± 0.036 |
5.805 ± 0.053 | 11.04 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.413 ± 0.026 | 4.867 ± 0.039 |
4.115 ± 0.026 | 4.67 ± 0.04 |
4.25 ± 0.037 | 6.763 ± 0.039 |
5.439 ± 0.04 | 6.142 ± 0.047 |
1.095 ± 0.017 | 3.423 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
